Research Articles
Beyond the Price Tag: A Comprehensive Analysis of NGS Cost-Effectiveness in Modern Chemogenomics
This article provides a rigorous, evidence-based assessment of the cost-effectiveness of Next-Generation Sequencing (NGS) against traditional single-gene testing methods in chemogenomics and drug development.
NGS vs Microarray for Chemical Perturbation Profiling: A 2025 Strategic Guide
Choosing between Next-Generation Sequencing (NGS) and microarrays for chemical perturbation studies is a critical decision that impacts data quality, cost, and biological insights.
A Practical Framework for Orthogonal Validation of NGS-Derived Chemogenomic Signatures in Drug Discovery
This article provides a comprehensive roadmap for researchers and drug development professionals to rigorously validate next-generation sequencing (NGS)-derived chemogenomic signatures.
Illumina vs. Nanopore Sequencing: A Strategic Comparison for Modern Chemogenomics and Drug Discovery
This article provides a comprehensive comparison of Illumina and Oxford Nanopore Technologies (ONT) sequencing platforms for chemogenomic applications.
NGS vs qPCR for Gene Expression Validation: A Strategic Guide for Chemogenomics Research
This article provides a comprehensive comparison of Next-Generation Sequencing (NGS) and quantitative PCR (qPCR) for gene expression validation in chemogenomics and drug development.
Addressing Sequencing Errors in Chemogenomic Variant Calling: Strategies for Robust Biomarker Discovery
Accurate variant calling is foundational for discovering genetic biomarkers of drug response in chemogenomics.
Taming the Data Deluge: Computational Strategies for Large-Scale Chemogenomic NGS
The integration of next-generation sequencing (NGS) into chemogenomics—the study of how genes influence drug response—generates datasets of immense scale and complexity, creating significant computational bottlenecks.
Automating NGS Workflows: A Strategic Guide to Unlocking Reproducible Chemogenomic Research
Next-generation sequencing (NGS) is revolutionizing drug discovery and biomedical research, but its potential is often limited by manual workflow inconsistencies.
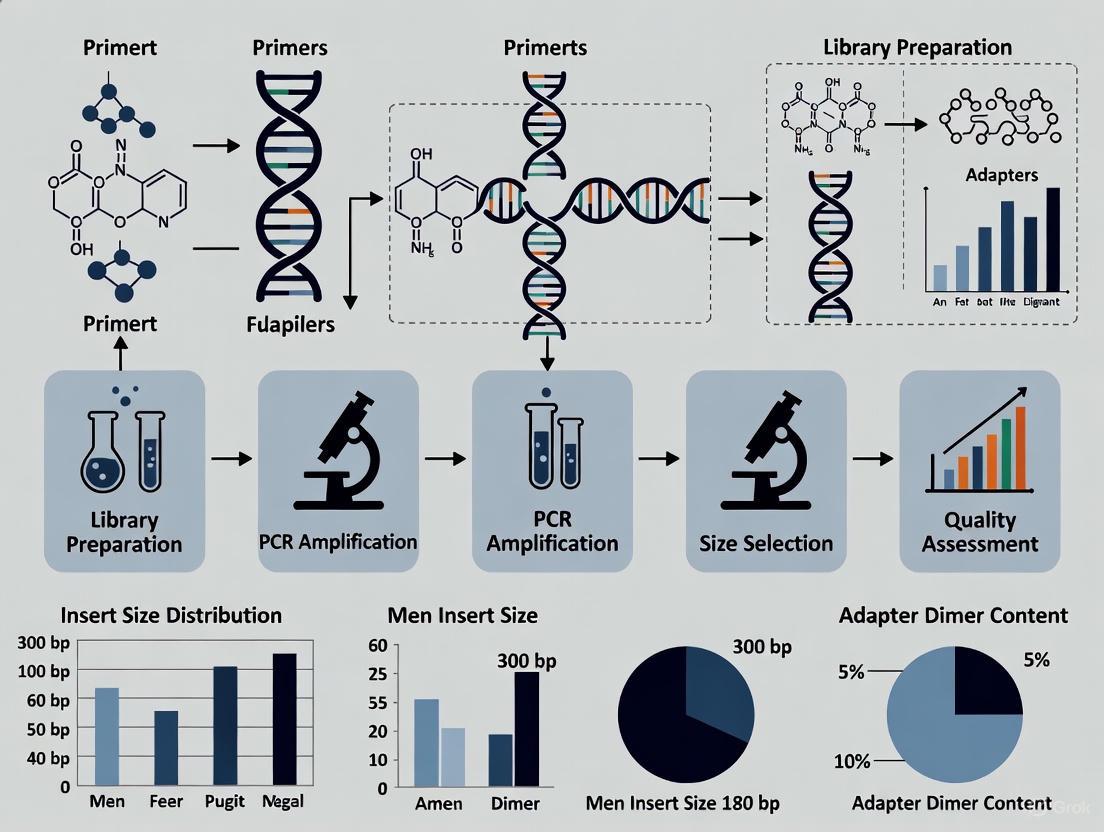
Ensuring Data Integrity: A Comprehensive Guide to Quality Control for Chemogenomic NGS Libraries
This article provides a comprehensive framework for implementing robust quality control (QC) protocols in chemogenomic Next-Generation Sequencing (NGS) workflows.
Breaking the Bottleneck: Strategies to Overcome NGS Data Analysis Challenges in Chemogenomics
Next-generation sequencing (NGS) has become indispensable in chemogenomics for uncovering the genetic basis of drug response and toxicity.