Overexpression vs Knockdown Screens: A Comprehensive Guide for Functional Genomics and Drug Discovery
This article provides a systematic comparison of overexpression (gain-of-function) and knockdown (loss-of-function) genetic screens, two pivotal methodologies in functional genomics.
Overexpression vs Knockdown Screens: A Comprehensive Guide for Functional Genomics and Drug Discovery
Abstract
This article provides a systematic comparison of overexpression (gain-of-function) and knockdown (loss-of-function) genetic screens, two pivotal methodologies in functional genomics. Tailored for researchers, scientists, and drug development professionals, we explore the foundational principles, distinct applications, and complementary strengths of each approach. The content delves into modern implementation using CRISPR activation (CRISPRa) and interference (CRISPRi) technologies, addresses common troubleshooting and optimization strategies, and presents a framework for rigorous validation and comparative analysis. By synthesizing insights from current literature and real-world case studies in oncology and neuroscience, this guide aims to inform strategic decisions in experimental design to deconvolute disease mechanisms and identify novel therapeutic targets.
Core Concepts: Unraveling the Principles of Gain-of-Function and Loss-of-Function Screens
Core Concepts and Mechanisms at a Glance
In functional genomics and drug development, overexpression and knockdown represent two foundational, complementary paradigms for probing gene function. Overexpression is primarily used to study gain-of-function (GOF) effects, while knockdown is used to study loss-of-function (LOF) effects [1]. The table below summarizes their core characteristics.
Table 1: Fundamental Comparison of Overexpression and Knockdown Paradigms
| Feature | Overexpression (GOF) | Knockdown (LOF) |
|---|---|---|
| Primary Objective | Introduce a functional gene to assess the effects of its enhanced activity [2]. | Reduce expression of an endogenous gene to study the consequences of its depletion [3]. |
| Molecular Mechanism | Increased protein levels leading to heightened or novel biological activity [4] [1]. | Reduced mRNA or protein levels, diminishing the gene's native function [3] [5]. |
| Typical Applications | • Identifying therapeutic genes for cell reprogramming [6]• Studying oncogene function [4]• Differentiation therapy [2] | • Validating essential genes for survival [3]• Modeling haploinsufficiency diseases [1]• Synthetic lethality screens [6] |
| Key Outcome | Emergence of new phenotypes or enhanced cellular processes [2] [4]. | Disruption of normal cellular functions, revealing gene necessity [3] [5]. |
Experimental Data and Performance Comparison
Empirical data from recent studies highlight the distinct transcriptional outcomes and phenotypic consequences of these two approaches.
Table 2: Quantitative Comparison from Experimental Studies
| Study Context | Perturbation Method | Key Quantitative Findings | Phenotypic Outcome |
|---|---|---|---|
| Mouse Retina Regeneration [7] | Cyclin D1 Overexpression & p27Kip1 Knockdown | • p27Kip1 knockdown alone: minimal MG proliferation• Cyclin D1 overexpression alone: 3-fold increase in MG proliferation vs. p27Kip1 knockdown• Combined approach: 5-fold increase in MG proliferation vs. cyclin D1 alone | Robust, self-limiting Müller glia (MG) proliferation, enabling potential neuron regeneration. |
| Hepatocellular Carcinoma (HCC) [2] | KDM6A Overexpression | Significant alteration in proliferation rate, cell cycle pattern, colony formation, and migration capacity of Huh-7 cells. | Attenuation of cancerous features, promotion of hepatocytic differentiation. |
| Cell Adhesion Study [3] | CRISPR-Cas9 Knockout & RNAi Knockdown | • Antibody transfection & CRISPR-Cas9: induced fewer deregulated mRNAs than RNAi• siRNAs: only 10% overlap of deregulated transcripts with negative controls | Distinct temporal onset dynamics for each method; antibodies induced phenotypic changes without altering target expression. |
| Amyotrophic Lateral Sclerosis (ALS) [5] | TDP-43 Loss-of-Function | Identification of 227 cryptic alternative polyadenylation (APA) events upon TDP-43 knockdown. | Nuclear depletion-induced cryptic APA, contributing to neurodegenerative disease pathology. |
Essential Research Reagent Solutions
The execution of overexpression and knockdown experiments relies on a suite of specialized molecular tools.
Table 3: Key Reagents for Genetic Perturbation Experiments
| Reagent / Method | Primary Function | Key Characteristics |
|---|---|---|
| Adeno-Associated Virus (AAV) [7] | In vivo gene delivery for overexpression or knockdown. | High transduction efficiency, cell-type-specific promoters (e.g., GFAP), broad tropism with different serotypes. |
| Lentiviral Vectors [2] | Stable integration of transgenes (e.g., KDM6A) or shRNAs into the host genome. | Enables long-term, stable gene expression or repression; suitable for difficult-to-transfect cells. |
| CRISPR Interference (CRISPRi) [8] [9] | Targeted gene knockdown using a catalytically dead Cas9 (dCas9) fused to repressor domains. | High specificity, reversible repression, minimal off-target effects compared to RNAi; uses dCas9-SALL1-SDS3 or dCas9-KRAB. |
| Synthetic sgRNA [9] | Chemically synthesized guide RNA for CRISPRi/CRISPRa applications. | Rapid delivery (transfection/electroporation), gene repression evident within 24 hours, enables easy multiplexing. |
| RNA Interference (RNAi) [3] | Sequence-specific degradation of mRNA using small interfering RNA (siRNA). | Rapid knockdown, but can have moderate off-target effects due to nonspecific mRNA binding. |
Detailed Experimental Protocols
Protocol 1: AAV-Mediated Overexpression and KnockdownIn Vivo
This protocol, based on retinal regeneration research [7], demonstrates a dual-vector strategy for manipulating gene expression in vivo.
- Vector Design: Clone the gene of interest (e.g., cyclin D1) or short hairpin RNA (shRNA) against the target (e.g., p27Kip1) into an AAV vector. Use a cell-type-specific promoter (e.g., GFAP for Müller glia) to restrict expression.
- Virus Production and Purification: Package the recombinant genome into AAV capsids (e.g., serotype 7m8 for retinal studies) using a standard transfection method in HEK293T cells. Purify the virus via ultracentrifugation.
- In Vivo Injection: Anesthetize the animal (e.g., postnatal day 6 mice) and perform intravitreal injection of the AAV vector into the eye.
- Phenotype Tracking: Administer a pulse of a nucleotide analog like EdU to label proliferating cells. After a predetermined period, harvest the tissue for analysis.
- Validation and Analysis:
- Immunohistochemistry: Analyze tissue sections for EdU incorporation and cell-type-specific markers (e.g., Sox9 for Müller glia) to quantify proliferation.
- qPCR: Quantify the level of target gene knockdown and/or transgene overexpression from harvested tissue.
Protocol 2: Lentiviral Overexpression in Cell Lines
This protocol, used to study tumor suppressors in hepatocellular carcinoma [2], outlines the process for stable gene overexpression in cultured cells.
- Lentiviral Vector Construction: Amplify the coding sequence (CDS) of the target gene (e.g., KDM6A) and clone it into a lentiviral expression plasmid (e.g., pLenti) using Gibson assembly. Verify the sequence.
- Lentivirus Production: Co-transfect HEK293T cells with the transfer plasmid (pLenti-KDM6A) and packaging plasmids (psPAX2 and pMD2.G) using a lipofectamine reagent.
- Virus Harvest and Titration: Collect the viral supernatant 48 hours post-transfection, filter it through a 0.45 μm membrane, and concentrate if necessary. Determine the viral titer using a suitable cell line (e.g., HT-1080).
- Cell Transduction: Seed the target cells (e.g., Huh-7) and transduce them with the viral supernatant in the presence of polybrene to enhance infection efficiency.
- Selection and Expansion: Select transduced cells using an antibiotic (e.g., puromycin) for 4-10 days. Expand the resistant population for downstream experiments.
- Phenotypic Assessment:
- Functional Assays: Perform proliferation assays (MTT), migration assays (scratch/wound healing), and colony formation assays.
- Molecular Analysis: Use qRT-PCR and Western blot to confirm gene expression and analyze downstream markers (e.g., EMT markers, differentiation genes).
Signaling Pathway and Workflow Visualizations
Diagram 1: Core Mechanistic Pathways of GOF and LOF
Diagram 2: Experimental Workflow for Combined GOF/LOF Study
The journey from cDNA libraries to modern CRISPR tools represents one of the most significant technological evolution in molecular biology. This transition has fundamentally transformed how researchers investigate gene function, moving from broad observational approaches to precise, systematic genetic manipulation. Within the context of comparing overexpression versus knockdown screening methodologies, this evolution highlights complementary approaches for understanding gene function—either by reducing gene expression to infer function or by enhancing expression to observe gain-of-function effects. This guide objectively compares these approaches through their historical development and current applications.
From cDNA to CRISPR: A Technical Revolution
The development of genetic screening tools has progressed through several distinct phases, each building upon the limitations of previous technologies. cDNA libraries, collections of complementary DNA sequences synthesized from messenger RNA, were among the first tools for gene discovery and expression studies. While revolutionary for their time, they lacked the precision for systematic functional genetics.
The emergence of RNA interference (RNAi) technologies marked a significant advancement, enabling targeted gene knockdown through introduction of small interfering RNAs. However, RNAi suffered from off-target effects and incomplete knockdown, limiting its reliability for large-scale screens [10].
The CRISPR-Cas9 system, adapted from a bacterial immune mechanism, represents the current state-of-the-art. Its precision, efficiency, and programmability have made it the preferred tool for both small-scale investigations and genome-wide screens [11]. The technology continues to evolve with base editing, prime editing, and activation/inhibition systems expanding its capabilities [12].
Comparative Analysis of Screening Approaches
Modern genetic screening employs two primary complementary approaches: loss-of-function (knockdown/knockout) and gain-of-function (overexpression) studies. The table below summarizes their key characteristics:
| Feature | Knockdown/Knockout Screens | Overexpression Screens |
|---|---|---|
| Mechanism | Utilizes CRISPR knockout (CRISPRn) or inhibition (CRISPRi) to disrupt or reduce gene function [12] [13] | Employs CRISPR activation (CRISPRa) or cDNA overexpression to enhance gene expression [13] |
| Primary Application | Identifying essential genes, drug targets, and genes whose loss confers resistance [13] | Discovering genes that drive phenotypes when overexpressed, including drug resistance mechanisms [13] |
| Typical Library Size | Varies from minimal (3-6 gRNAs/gene) to comprehensive (10+ gRNAs/gene) [14] | Similar range to knockout libraries, with 4-10 gRNAs per gene common [13] |
| Key Advantages | Reveals gene essentiality; identifies synthetic lethal interactions | Uncovers oncogenes and resistance mechanisms; complements knockout findings [13] |
| Limitations | May miss genes requiring overexpression to reveal function | Can produce non-physiological effects; may activate compensatory mechanisms |
Table 1: Comparison of knockdown/knockout versus overexpression screening approaches
Performance Benchmarking: Quantitative Data
Recent benchmarking studies provide quantitative comparisons of CRISPR library performance. The table below summarizes key metrics from empirical evaluations:
| Library Name | gRNAs per Gene | Essential Gene Depletion | Non-essential Enrichment | Best Application Context |
|---|---|---|---|---|
| Vienna-single | 3 | Strongest depletion | Minimal enrichment | Minimal library size applications [14] |
| Yusa v3 | 6 | Moderate depletion | Moderate enrichment | Standard screening conditions [14] |
| Croatan | 10 | Strong depletion | Low enrichment | High-sensitivity essentiality screens [14] |
| Vienna-dual | 3 pairs | Strongest depletion | Some fitness reduction observed | Maximum sensitivity screens [14] |
| Brunello | 4 | Strong depletion | Low enrichment | General purpose knockout screens [13] |
Table 2: Performance benchmarking of commonly used CRISPR libraries
Notably, recent studies demonstrate that minimal libraries with only 3 highly efficient guides per gene can perform as well or better than larger libraries when guides are selected using principled criteria like VBC scores [14]. Dual-targeting libraries, where two sgRNAs target the same gene simultaneously, show enhanced depletion of essential genes but may induce a mild fitness cost even in non-essential genes, possibly due to increased DNA damage response [14].
Experimental Protocols: From Library Design to Analysis
CRISPR Knockout Screen Protocol
The following workflow details a typical genome-wide CRISPR knockout screen, based on established methodologies [13]:
- Library Selection: Choose an appropriate library (e.g., Brunello with 4 gRNAs per gene targeting 19,114 genes) [13]
- Lentiviral Production: Package the sgRNA library into lentiviral particles in HEK293T cells
- Cell Transduction: Transduce target cells at low MOI (~0.1) to ensure single integration events
- Selection: Apply puromycin selection (1 μg/mL) for 3-7 days to eliminate untransduced cells
- Screening: Split cells into treatment and control groups; for ATR inhibitor screens, use high doses (e.g., 1.5μM VE822) that kill ~90% of cells over 108 hours [13]
- Genomic DNA Extraction: Harvest surviving cells and extract genomic DNA after 10-14 days
- sgRNA Amplification: PCR-amplify sgRNA regions using barcoded primers for multiplexing
- Sequencing: Perform Illumina sequencing to quantify sgRNA abundance
- Bioinformatic Analysis: Use MAGeCK or RSA algorithms to identify significantly enriched/depleted sgRNAs [13]
CRISPR Activation Screen Protocol
CRISPR activation screens follow a similar workflow with key modifications [13]:
- Library Selection: Employ a CRISPR activation library (e.g., Calabrese library)
- dCas9-VPR Expression: Use cells stably expressing the dCas9-VPR activation system
- sgRNA Delivery: Transduce with activation library as described above
- Selection and Screening: Follow similar selection and treatment steps as knockout screens
- Analysis: Identify genes whose overexpression confers resistance or other phenotypes
Library Customization Protocol
Customizing existing libraries using CRISPR/Cas9 itself provides a efficient alternative to building new libraries [15]:
- Identify Target gRNAs: Select gRNAs to eliminate from existing library
- Design rc-gRNAs: Create reverse-complementary gRNAs targeting undesired gRNAs in the library plasmid DNA
- Form RNPs: Complex purified Cas9 proteins with rc-gRNAs to form ribonucleoproteins
- Library Treatment: Incubate pooled CRISPR library plasmid DNA with RNPs (e.g., 20μg library DNA with 10μg Cas9 + 7.5μg rc-gRNAs for 8h at 37°C) [15]
- Purify Depleted Library: Recover processed library DNA through isopropanol precipitation
- Quality Control: Verify specific gRNA depletion via qPCR and deep sequencing
Visualizing Screening Approaches and Workflows
Knockdown vs. Overexpression: Conceptual Framework
Integrated Screening Workflow
The Scientist's Toolkit: Essential Research Reagents
| Reagent/Library | Type | Primary Application | Key Features |
|---|---|---|---|
| Brunello Library [13] | CRISPR knockout | Genome-wide loss-of-function screens | 76,441 gRNAs targeting 19,114 genes; 4 gRNAs/gene |
| Calabrese Library | CRISPR activation | Genome-wide gain-of-function screens | dCas9-VPR system; targeted gene overexpression |
| GeCKO v2 | CRISPR knockout | Customizable screening | Dual-sgRNA libraries; modular design |
| Vienna Library | CRISPR knockout/activation | Minimal library screens | Top 3 VBC-scored gRNAs per gene; high efficiency [14] |
| ATR Inhibitors (VE822, AZD6738) [13] | Small molecule compounds | DNA damage response studies | Selective ATR kinase inhibition; clinical relevance |
| MAGeCK | Bioinformatics tool | CRISPR screen analysis | Identifies positively/negatively selected genes |
| Chronos | Bioinformatics tool | Time-series screen analysis | Models gene fitness across multiple timepoints [14] |
Table 3: Essential research reagents for genetic screens
The evolution from cDNA libraries to modern CRISPR tools has provided researchers with an unprecedented ability to systematically probe gene function. The choice between knockdown and overexpression approaches depends heavily on the biological question:
- Knockdown/knockout screens excel at identifying essential genes and synthetic lethal interactions, particularly for investigating drug sensitivity [13].
- Overexpression/activation screens powerfully reveal oncogenes, resistance mechanisms, and genes that produce phenotypes only when upregulated [13].
- Dual screening approaches, combining both methods, provide the most comprehensive understanding of gene function in biological processes and therapeutic contexts [13].
Recent advances in library design, particularly minimal libraries with highly efficient guides, have made genome-wide screens more accessible and cost-effective [14]. Furthermore, AI tools like CRISPR-GPT are now accelerating experimental design and making CRISPR technologies accessible to non-specialists [16]. As these technologies continue to converge, they promise to further accelerate the pace of functional genomics and therapeutic discovery.
| Reagent Type | Specific Examples | Key Function in Experiments |
|---|---|---|
| CRISPRi Effectors | dCas9-KRAB, dCas9-SALL1-SDS3, dCas9-ZIM3(KRAB)-MeCP2(t) [9] [17] [18] | Engineered fusion proteins that bind DNA without cutting and recruit repressive complexes to silence target genes. |
| CRISPRa Effectors | dCas9-VP64, dCas9-VPR, SAM system (dCas9-VP64-MS2-P65-HSF1) [18] [19] [20] | Fusion proteins or complexes that recruit transcriptional activators to the promoter region to upregulate gene expression. |
| Guide RNAs | Synthetic sgRNA, crRNA:tracrRNA duplex, Lentiviral sgRNA [9] [19] | Programmable RNA components that direct the dCas9 effector to specific genomic loci based on sequence complementarity. |
| Delivery Vehicles | Lentiviral particles, Transient mRNA, Plasmids, Baculovirus (for ASCs) [9] [19] [20] | Methods for introducing CRISPR components into cells; chosen based on desired duration (transient vs. stable) and cell type. |
| Controls | Non-targeting sgRNA, Targeting sgRNA for housekeeping genes (e.g., PPIB) [9] [21] | Essential reagents to account for non-specific effects of delivery and the CRISPR machinery itself. |
CRISPRa/i, RNAi, and cDNA Overexpression: A Comparative Guide
In the functional genomics toolkit, researchers have multiple methods to perturb gene expression and investigate gene function. The key strategic choice often lies between gain-of-function (GOF) and loss-of-function (LOF) approaches [22]. cDNA overexpression is a classic GOF technique, while CRISPR interference (CRISPRi) and RNA interference (RNAi) are two central LOF methods [23] [22]. This guide objectively compares the performance, specificity, and applications of these key technological platforms—CRISPRi, RNAi, and cDNA overexpression—framed within the context of overexpression versus knockdown screens for target discovery and validation.
The core distinction between these technologies lies in their mechanism and level of action: CRISPRi acts at the genomic DNA level to prevent transcription, RNAi operates at the mRNA level to trigger transcript degradation, and cDNA overexpression introduces an exogenous transcript to augment gene function [18] [21] [22].
The following table provides a detailed, data-driven comparison of their key characteristics.
| Feature | CRISPRi | RNAi | cDNA Overexpression |
|---|---|---|---|
| Mechanism of Action | dCas9-repressor fusion binds DNA and blocks transcription [9] [18]. | siRNA/shRNA binds mRNA, leading to its degradation [21]. | Introduces exogenous cDNA copy of the gene to increase expression [22]. |
| Level of Intervention | Transcriptional (DNA level) [18]. | Post-transcriptional (mRNA level) [21]. | Transcriptional (via exogenous promoter) [22]. |
| Reversibility | Reversible (knockdown) [18] [22]. | Reversible (knockdown) [23]. | Reversible (dependent on delivery method). |
| Genetic Alteration | Epigenetic/modulation, no DNA cleavage [9] [18]. | None (targets mRNA) [21]. | Addition of genetic material. |
| Typical Knockdown Efficiency | Robust (down to 20-30% of baseline) [9]. | Variable, can be high [21]. | Not Applicable (Overexpression) |
| Duration of Effect | Extended (days to weeks) [9]. | Transient (typically 3-7 days) [9]. | Variable (transient to stable). |
| Multiplexing Capacity | High (easy to pool multiple sgRNAs) [9] [22]. | Moderate (can be limited by competition) [9]. | Low (limited by vector capacity and promoter compatibility). |
| Major Source of Off-Target Effects | Off-target DNA binding [21]. | Seed sequence homology with non-target mRNAs [21]. | Ectopic, non-physiological expression levels. |
| Key Advantages | High specificity; reversible; gentle knockdown; excellent for non-coding genes [9] [18] [22]. | Rapid deployment; well-established [21]. | Studies dominant-positive effects; expresses mutated genes; complements knockdown studies [10]. |
Supporting Experimental Data and Performance
Independent, direct comparisons and functional assays highlight critical performance differences between these methods.
- Efficiency and Duration: In head-to-head tests, CRISPRi-mediated gene repression is evident 24 hours after transfection and persists through at least 96 hours, with maximal repression at 48-72 hours [9]. RNAi effects are typically transient, often lasting a few days, which can be a limitation for long-term studies [9].
- Specificity and Off-Target Effects: A systematic study comparing off-target effects at the whole-transcriptome level found that all three major LOF methods (RNAi, LNA gapmers, and CRISPRi) yielded non-negligible off-target effects [21]. However, the nature of these effects differs. RNAi off-targets are largely driven by seed sequence homology in the siRNA, leading to unintended mRNA degradation [21]. In contrast, CRISPRi introduced in a polyclonal population showed limited sequence-dependent off-target effects, though single-cell clones from this population could exhibit unique and reproducible transcriptional signatures [21].
- Multiplexing and Screening Performance: CRISPRi and CRISPRa are highly effective for pooled genetic screens. CRISPRi knockdown phenotypes robustly identify essential genes, while CRISPRa can reveal genes whose overexpression is detrimental, such as tumor suppressors [22]. A key advantage of CRISPRi synthetic sgRNA is the straightforward pooling of guides to enhance knockdown of a single gene or to enable the simultaneous knockdown of multiple genes without substantial loss of efficiency, a process known as multiplexing [9].
Detailed Experimental Protocols
To ensure reproducibility, below are detailed methodologies for implementing these technologies, as cited in the literature.
CRISPRi Knockdown with Synthetic sgRNA
This protocol is adapted from Horizon Discovery's CRISPRi application guide [9].
- Cell Line: U2OS cells stably expressing integrated dCas9-SALL1-SDS3 or dCas9-KRAB.
- Transfection:
- Plate cells at 10,000 cells/well in a multi-well plate.
- Transfect with a pool of pre-designed synthetic sgRNAs (total concentration 25 nM) using DharmaFECT 4 Transfection Reagent.
- Include a non-targeting control (NTC) sgRNA.
- Harvest and Analysis:
- Harvest cells at 72 hours post-transfection for maximal repression.
- Isolate total RNA and synthesize cDNA.
- Measure relative gene expression using RT-qPCR with the ∆∆Cq method, using GAPDH or ACTB as a housekeeping gene.
RNAi Knockdown with siRNA
This protocol is based on the methods used in the ADAM28 prostate cancer study [24].
- Cell Line: Human prostate cancer cells (e.g., LNCaP, DU145).
- Transfection:
- Seed cells into 12-well culture dishes.
- Transfert with 10 nM Silencer Select siRNA (e.g., targeting ADAM28) using X-tremeGENE HP DNA transfection reagent.
- Use a scrambled siRNA sequence as a negative control.
- Incubate for 48 hours in growth media free of penicillin/streptomycin.
- Functional Assays:
- Assess proliferation using assays like MTT or BrdU.
- Assess migration using Transwell or wound-healing assays.
- Confirm knockdown via western blotting or RT-qPCR.
cDNA Overexpression
This protocol is illustrative of the approach used in GOF studies, as seen in the ADAM28 study [24].
- Cell Line: Relevant cell model (e.g., RWPE1 normal prostate epithelial cells).
- Transfection:
- Transfect cells with plasmid vectors (e.g., pCMV Tag4A ADAM28) using a suitable transfection reagent.
- Use an empty vector (e.g., pCMV Tag4A) as a control.
- Validation and Functional Assays:
- Confirm overexpression 48 hours post-transfection using immunocytochemistry or western blotting.
- Perform functional assays (proliferation, migration) paralleling the knockdown experiments.
Visualization of Workflows and Mechanisms
The following diagrams illustrate the core mechanisms and experimental workflows for these technologies.
Experimental Workflow for Functional Screening
In functional genomics, the systematic manipulation of gene expression levels—through either overexpression or knockdown—serves as a fundamental strategy for elucidating gene function. The core premise is that observing phenotypic consequences resulting from too much or too little of a gene product can reveal its normal biological role within a cell or organism [25]. Overexpression studies introduce an excess of a gene's product, often revealing its potential roles in driving cellular processes and potentially uncovering dominant-negative or neomorphic effects. Conversely, knockdown techniques reduce gene expression, mimicking loss-of-function mutations and highlighting genes essential for specific biological processes [26]. Together, these complementary approaches form a powerful toolkit for deconstructing complex biological systems, defining signaling pathways, and identifying potential therapeutic targets in disease contexts such as cancer [27] [13] [28].
Experimental Approaches for Modulating Gene Expression
Gene Knockdown Methodologies
Gene knockdown refers to experimental techniques that reduce the expression of one or more genes. This can be achieved through methods that do not permanently alter the chromosomal DNA, leading to a transient knockdown, or through genetic modification, resulting in a stable "knockdown organism" [25].
- RNA Interference (RNAi): This is a widely used knockdown method that utilizes double-stranded RNA to induce sequence-specific gene silencing. The process involves introducing small double-stranded interfering RNAs (siRNAs) into the cytoplasm. These siRNAs are loaded into the RNA-induced silencing complex (RISC), which uses them as a template to locate and cleave complementary messenger RNA (mRNA) transcripts, preventing their translation into protein [25] [29]. For stable, long-term knockdown, short hairpin RNAs (shRNAs) can be expressed from viral or plasmid vectors, which are then processed into siRNAs inside the cell [29].
- CRISPR Interference (CRISPRi): While the CRISPR-Cas system is famously used for gene knockout, it can also be engineered for knockdown. Using a catalytically "dead" Cas9 (dCas9) fused to repressive domains, the system can be targeted to specific gene promoters to block transcription without cutting the DNA, effectively reducing gene expression [13] [25].
- Morpholino Oligonucleotides: These are antisense oligonucleotides used primarily in developmental biology models like zebrafish. They are designed to bind to specific mRNA sequences, blocking translation or pre-mRNA splicing. A key advantage is their stability, as they are not degraded by cellular enzymes, but their effect is diluted with each cell division [26].
Gene Overexpression Methodologies
Overexpression studies aim to increase the production of a specific gene product to observe the resulting phenotypic effects.
- Recombinant Expression Vectors: This common method involves cloning the cDNA of a target gene into a plasmid vector under the control of a strong promoter, such as the CAG promoter (a hybrid of CMV enhancer and chicken beta-actin promoter) [30]. The recombinant plasmid is then transfected into cells, leading to the production of the target gene's mRNA and protein at levels significantly higher than normal [31].
- cDNA Screening: In this approach, libraries containing hundreds of cDNAs are systematically transfected into cell lines. This allows for the high-throughput identification of genes that, when overexpressed, modulate specific pathways of interest, such as the Sonic hedgehog (SHH) signaling pathway [28].
- CRISPR Activation (CRISPRa): This gain-of-function version of CRISPR uses a dCas9 protein fused to transcriptional activators. When guided to a gene's promoter, the complex can significantly upregulate the gene's transcription, providing a powerful tool for genome-wide overexpression screens [13].
Advanced Screening Platforms
Modern genetic screens often employ dual approaches to comprehensively map gene function. A prime example is the use of dual genome-wide CRISPR knockout and CRISPR activation screens to identify genes that confer resistance or sensitivity to drugs like ATR inhibitors. This dual strategy allows researchers to simultaneously probe loss-of-function and gain-of-function phenotypes in a single, systematic experiment [13].
Furthermore, innovative tools like the "Double UP" plasmid address technical challenges in functional studies. This dual-fluorescent plasmid uses a LoxP-flanked cassette and limiting Cre recombinase to generate an internal control within the same population of transfected cells. This allows for a direct and robust comparison between control (e.g., mNeonGreen-positive) and experimentally manipulated (e.g., mScarlet-positive) cells, significantly reducing variability and improving the reliability of conclusions drawn from overexpression or knockdown experiments [30].
Comparative Analysis: Overexpression vs. Knockdown Screens
The following table summarizes the core characteristics, applications, and outputs of overexpression and knockdown screens, illustrating their complementary nature in functional genomics.
Table 1: Comparative overview of overexpression and knockdown screens.
| Feature | Overexpression Screens | Knockdown Screens |
|---|---|---|
| Primary Goal | Identify genes that drive processes or confer capabilities when overexpressed [13] [28]. | Identify genes essential for specific biological processes or cell viability when suppressed [13] [25]. |
| Typical Approach | cDNA libraries, CRISPR activation (CRISPRa) [13] [28]. | RNAi (siRNA/shRNA), CRISPR knockout [13] [25]. |
| Key Readouts | Enhanced proliferation, migration, drug resistance, pathway activation [27] [28] [31]. | Impaired proliferation, migration, increased apoptosis, pathway inhibition, sensitization to drugs [27] [13]. |
| Major Utility | Uncovering oncogenes, signaling pathway modulators, and mechanisms of drug resistance [13] [28]. | Identifying tumor suppressors, essential genes, and synthetic lethal interactions for therapy [27] [13]. |
Case Studies in Cancer Research
Targeting the P2X7 Receptor in Non-Small Cell Lung Cancer
A clear example of how both overexpression and knockdown validate a therapeutic target comes from research on the P2X7 receptor in non-small cell lung cancer (NSCLC). Researchers constructed recombinant plasmids to overexpress or knock down the P2X7 receptor in LLC and LA795 lung cancer cells.
- Overexpression Effects: Forcing the expression of the P2X7 receptor promoted the migration and invasion of lung cancer cells. It also activated pro-tumorigenic signaling pathways, including PI3K/Akt/GSK-3β and JNK, and induced changes associated with epithelial-mesenchymal transition (EMT) [27].
- Knockdown Effects: Reducing P2X7 receptor expression produced the opposite effects: it suppressed cell proliferation, promoted apoptosis, and inhibited tumor growth in both cell-based and animal models. The expression levels of PI3K/Akt/GSK-3β, JNK, and EMT markers were consistently downregulated, confirming the pathway involvement [27].
This dual-approach study conclusively demonstrated that downregulating the P2X7 receptor effectively suppresses tumor growth and progression, establishing its potential as a therapeutic target for NSCLC [27].
Table 2: Quantitative effects of P2X7 receptor modulation in lung cancer models (based on [27]).
| Experimental Group | Proliferation Impact | Migration/Invasion Impact | Key Signaling Pathway Modulation |
|---|---|---|---|
| P2X7 Overexpression | Promoted growth | Increased migration and invasion | Activation of PI3K/Akt/GSK-3β and JNK; induction of EMT |
| P2X7 Knockdown | Suppressed growth; promoted apoptosis | Inhibited migration and invasion | Suppression of PI3K/Akt/GSK-3β and JNK; inhibition of EMT |
Identifying Modulators of ATR Inhibitor Resistance
The power of dual CRISPR screens is exemplified in research aimed at understanding resistance to ATR inhibitors (ATRi), emerging cancer therapeutics. A study performed both genome-wide CRISPR knockout and CRISPR activation screens in HeLa and MCF10A cells treated with two different ATR inhibitors, VE822 and AZD6738 [13].
- Knockdown Screen Findings: The knockout screen identified genes that, when lost, made cells more resistant to ATR inhibitors. The top hits were enriched for biological pathways like protein translation, DNA replication, and sister chromatid cohesion [13].
- Overexpression Screen Findings: The parallel activation screen identified genes that, when overexpressed, could also confer resistance to ATRi.
- Integrated Analysis: By combining both screens, the study revealed that resistance mechanisms are varied. They can involve restoring DNA replication fork progression or preventing ATR inhibitor-induced apoptosis. For instance, the study described how MED12-mediated inhibition of the TGFβ pathway can regulate replication fork stability and survival upon ATR inhibition [13].
This comprehensive approach successfully cataloged genetic determinants of ATRi resistance, providing a foundation for personalized cancer medicine by identifying potential biomarkers to predict patient response [13].
Dissecting Sonic Hedgehog Signaling in Down Syndrome
Research into the link between trisomy 21 (Down syndrome) and cerebellar defects showcases the use of a targeted overexpression screen. Given that chromosome 21 does not encode any known canonical Sonic hedgehog (SHH) pathway components, researchers systematically overexpressed 163 chromosome 21 cDNAs in SHH-responsive mouse cell lines to find which ones modulated the pathway [28].
- Functional Discovery: The screen successfully identified specific chromosome 21 genes that either upregulated (e.g., DYRK1A) or inhibited (e.g., HMGN1) SHH signaling [28].
- Validation in Disease Models: The relevance of these findings was confirmed by analyzing gene expression in the cerebella of Ts65Dn and TcMAC21 mouse models of Down syndrome. Furthermore, overexpressing four prioritized genes (B3GALT5, ETS2, HMGN1, and MIS18A) in primary granule cell precursors inhibited their SHH-dependent proliferation, linking these genes to the cerebellar hypoplasia phenotype [28].
This study highlights how a focused overexpression screen can pinpoint dosage-sensitive genes that disrupt key developmental pathways, suggesting new therapeutic avenues for ameliorating associated phenotypes [28].
Essential Research Reagents and Tools
The experimental strategies discussed rely on a suite of core reagents and tools that enable precise genetic manipulation.
Table 3: Key research reagents and solutions for modulation of gene expression.
| Reagent / Tool | Function | Example Use Cases |
|---|---|---|
| siRNA / shRNA | Synthetic double-stranded RNAs for transient (siRNA) or stable (shRNA) gene knockdown via the RNAi pathway [25] [29]. | Knockdown of EpCAM in colorectal cancer cells to study migration [31]. |
| CRISPR-Cas9 System | A versatile system using a Cas9 nuclease and guide RNA (gRNA) for targeted gene knockout. Can be adapted for knockdown (CRISPRi) or activation (CRISPRa) [13] [25]. | Dual genome-wide screens for ATR inhibitor resistance genes [13]. |
| Morpholino Oligos | Stable antisense oligonucleotides that block mRNA translation or splicing; ideal for embryonic studies [26]. | Gene knockdown in zebrafish models for developmental biology [26]. |
| Plasmid Vectors | Engineered DNA constructs for delivering genes (for overexpression) or shRNA sequences (for knockdown) into cells [31] [30]. | pCDH-EpCAM for overexpression; pGMC-KO-EpCAM for knockdown [31]. |
| Transfection Reagents | Lipid-based or other reagents that facilitate the uptake of nucleic acids (siRNA, plasmids) into cultured cells [29]. | siLenFect for siRNA delivery into MIA PaCa-2 pancreatic cancer cells [29]. |
| The Double UP Plasmid | A dual-fluorescent plasmid that, when co-transfected with limiting Cre, generates internal control and experimental cell populations in a single culture [30]. | Controlling for variability in in utero electroporation experiments studying neuronal migration [30]. |
Visualizing the Core Concepts and Workflows
How Gene Dosage Reveals Function
This diagram illustrates the fundamental biological rationale that underpins both overexpression and knockdown studies. It shows how deviations from normal gene expression levels can lead to observable phenotypic changes, thereby revealing the gene's function.
Dual CRISPR Screening Workflow
This flowchart outlines the integrated experimental pipeline for conducting dual genome-wide CRISPR knockout and activation screens, as used to identify mechanisms of drug resistance.
Key Signaling Pathway in a Functional Study
This diagram summarizes a key signaling pathway identified through overexpression/knockdown studies, showing how a target gene (P2X7R) influences cancer hallmarks through specific molecular cascades.
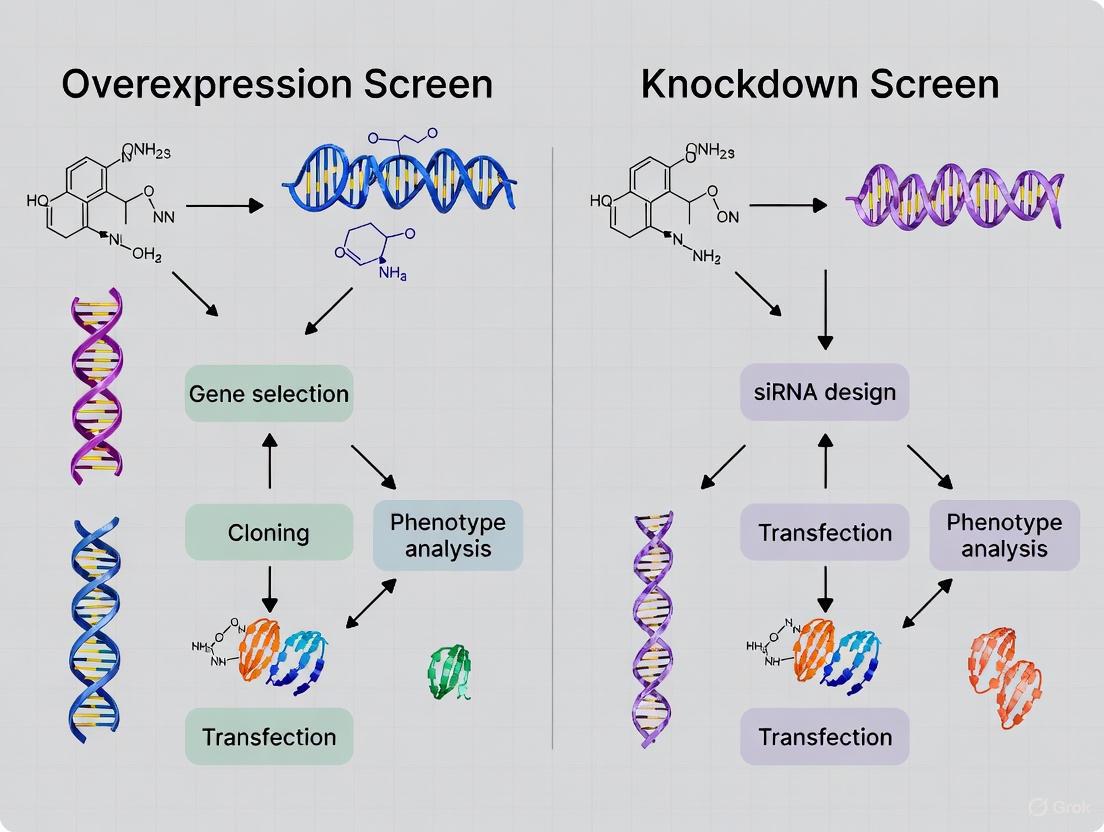
In functional genomics, researchers employ two powerful, complementary approaches to decipher gene function: overexpression and knockdown. Overexpression (or activation) screens investigate the consequences of increasing a gene's expression level, revealing insights into gene function when it is amplified or activated. Conversely, knockdown (or knockout) screens reduce or abolish gene expression to understand a gene's normal function by observing the phenotypic consequences of its loss. While often used independently, when applied in tandem, these strategies provide a more complete picture of gene function and its role in biological systems, from fundamental cellular processes to disease mechanisms and therapeutic development. This guide objectively compares the performance, applications, and experimental outcomes of these two foundational approaches in modern biological research.
Core Conceptual Frameworks and Biological Questions
Table 1: Fundamental Characteristics of Overexpression and Knockdown Approaches
| Feature | Overexpression/Activation | Knockdown/Knockout |
|---|---|---|
| Primary Goal | Identify genes whose increased activity induces a phenotype (e.g., drug resistance, pathogen restriction). | Identify genes whose loss induces a phenotype (e.g., cell death, sensitization to a drug). |
| Typical Screen Output | Hits that confer a selective advantage (e.g., survival, growth). | Hits that confer a selective disadvantage or synthetic lethality. |
| Key Biological Questions | What genes can drive a specific process or cellular state when activated? What genes can suppress a phenotype when overexpressed? | What genes are essential for a specific process or for cell survival? What genes enhance a phenotype when lost? |
| Common Technologies | cDNA libraries, ORF archives, CRISPR activation (CRISPRa). | RNAi (siRNA, shRNA), CRISPR knockout (CRISPRko). |
Experimental Designs and Methodologies
Key Experimental Protocols
The choice of experimental protocol is critical for the success and interpretation of both overexpression and knockdown screens.
Protocol 1: Dual Genome-Wide CRISPR Knockout and Activation Screening This protocol, as employed in a study investigating resistance to ATR inhibitors, leverages the power of CRISPR technology to simultaneously probe loss-of-function and gain-of-function phenotypes in a single, integrated workflow [13].
- 1. Library Design:
- 2. Cell Transduction: Transduce a population of cells (e.g., HeLa, MCF10A) with the lentiviral library at a low multiplicity of infection (MOI) to ensure most cells receive a single genetic perturbation [13].
- 3. Selection Pressure: Apply a selective pressure, such as a drug (e.g., ATR inhibitors VE822 or AZD6738). The dose and duration are critical; high doses can be used to specifically select for resistant clones [13].
- 4. Hit Identification: Harvest surviving cells, amplify integrated sgRNAs via PCR, and perform next-generation sequencing (NGS). Bioinformatics tools (e.g., the RSA algorithm) are used to identify sgRNAs, and thus genes, that are significantly enriched or depleted in the treated population compared to a control [13].
Protocol 2: Overexpression Screen Using a Defined Gene Library This methodology is used to identify individual genes that can induce a specific phenotype, such as resistance to pathogen infection [32].
- 1. Library Cloning: Curate a library of genes of interest (e.g., 414 interferon-stimulated genes) into a lentiviral expression vector that often co-expresses a fluorescent marker like tagRFP for tracking [32].
- 2. Cell Transduction: Transduce target cells (e.g., A549 lung epithelial cells) with the library. A "one-gene-per-well" format in a plate-based assay is common for such focused screens [32].
- 3. Phenotypic Assay: Challenge the transduced cells with the stimulus (e.g., GFP-expressing Toxoplasma gondii) [32].
- 4. Automated Analysis: Use high-throughput automated microscopy and image analysis to quantitatively measure the phenotypic outcome (e.g., size and number of parasitophorous vacuoles) [32].
- 5. Hit Validation: Identify genes that potently restrict the phenotype (e.g., infection) and validate them through secondary assays in multiple cell lines [32].
Research Reagent Solutions
Table 2: Essential Research Tools for Overexpression and Knockdown Studies
| Reagent / Solution | Function | Example Use Cases |
|---|---|---|
| CRISPR Knockout Library (e.g., Brunello) | A pooled library of sgRNAs for genome-wide loss-of-function screening [13]. | Identifying essential genes and drug sensitizers [13] [33]. |
| CRISPR Activation Library (e.g., Calabrese) | A pooled library of sgRNAs for targeted transcriptional activation of endogenous genes [13]. | Identifying genes that confer drug resistance or suppress phenotypes when overexpressed [13]. |
| ASKA Library (E. coli ORF Archive) | A complete set of E. coli open reading frames (ORFs) for overexpression screening [34]. | Genome-scale screening of beneficial upregulation targets in bacteria [34]. |
| Synthetic sRNAs (e.g., BHR-sRNA Platform) | Engineered small RNAs for targeted gene knockdown at the translational level in diverse bacteria [35]. | Knocking down virulence factors in pathogens or metabolic genes in industrial strains [35]. |
| Lentiviral Expression Vectors (e.g., pSUPER.retro) | Viral delivery systems for stable integration and expression of shRNAs or cDNAs in mammalian cells [36]. | Achieving stable, long-term gene knockdown or overexpression [36]. |
| Fluorescence-Activated Cell Sorting (FACS) | Technology to sort cells based on fluorescent markers, often used to isolate populations with desired phenotypes in screens [34]. | Enriching for cells with high production of a metabolite (e.g., free fatty acids) or specific surface markers [34]. |
Comparative Experimental Data and Outcomes
Direct comparisons and individual studies highlight the distinct yet complementary insights gained from overexpression and knockdown screens.
Table 3: Comparative Experimental Data from Overexpression and Knockdown Studies
| Study Context | Overexpression Findings | Knockdown Findings | Complementary Insight |
|---|---|---|---|
| Non-Small Cell Lung Cancer (NSCLC) [27] | Overexpression of the P2X7 receptor promoted migration, invasion, and tumor growth of NSCLC cells, involving PI3K/Akt/GSK-3β and JNK pathways. | Knockdown of the P2X7 receptor suppressed proliferation, migration, invasion, promoted apoptosis, and inhibited tumor growth. | The P2X7 receptor functions as a consistent oncogenic driver; its bidirectional modulation validates it as a high-confidence therapeutic target. |
| ATR Inhibitor Resistance [13] | A genome-wide CRISPRa screen identified genes that, when overexpressed, confer resistance to ATR inhibitors (e.g., via restoring replication fork progression). | A parallel CRISPRko screen identified genes whose loss confers resistance to ATR inhibitors (e.g., by preventing apoptosis). | Dual screens revealed varied resistance mechanisms, providing a more comprehensive landscape of potential biomarkers for therapy. |
| Free Fatty Acid Production in E. coli [34] | Overexpression of rfaY (involved in LPS biosynthesis) increased FFA production by 207.8% by enhancing membrane stability and homeostasis. |
(Inferred) Knockdown of competing pathway genes would be used to redirect metabolic flux, but was not the focus of this specific overexpression screen. | Highlights how overexpression can be used to identify non-obvious, non-metabolic gene targets (membrane integrity) for enhancing bioproduction. |
| Bacterial Gene Knockdown [35] | (Not Applicable) | A broad-host-range synthetic sRNA platform achieved >50% knockdown of target genes in 12 diverse bacterial species, mitigating virulence and optimizing metabolism. | Demonstrates the robustness and wide applicability of knockdown tools across diverse organisms, which is crucial for comparative biology and engineering. |
Integrated Workflow and Pathway Visualization
The following diagram illustrates the logical relationship and complementary nature of the two approaches within a typical functional genomics investigation, leading to a unified biological insight.
The mechanistic insights gained from these screens are often visualized through signaling pathways. The study on the P2X7 receptor in NSCLC provides a clear example of how a single target can be interrogated from both directions to validate its role and mechanism [27].
Overexpression and knockdown screens are not opposing strategies but are fundamentally complementary. As the experimental data demonstrates, knockdown screens excel at identifying genes that are essential for a biological process, revealing vulnerabilities and core components of pathways. Overexpression screens, in contrast, are powerful for discovering genes that are sufficient to drive a process, uncovering potential oncogenes, resistance mechanisms, and limiting factors in biosynthetic pathways.
The most powerful insights emerge when these approaches are used in tandem. The dual CRISPR screen on ATR inhibitor resistance is a prime example, where both methods identified resistance genes but through entirely different mechanisms [13]. Similarly, the bidirectional modulation of the P2X7 receptor provides a more robust and validated conclusion about its function in cancer than either approach could alone [27].
A critical consideration in interpreting these screens is the potential for non-specific or confounding effects. A notable analysis of LINCS L1000 data revealed that transcriptional profiles from knocking down and overexpressing the same gene were often positively correlated, contrary to the intuitive expectation of anticorrelation [10]. This highlights a "not a happy cell" effect, where perturbations can induce a general stress response, underscoring the necessity for careful validation through secondary assays, such as the CelFi assay for cellular fitness [33].
In conclusion, the strategic selection between overexpression and knockdown—or better yet, their integrated application—provides a powerful toolkit for deconstructing complex biological systems. For researchers in drug development, this combined approach is indispensable for target identification, understanding mechanism of action, and anticipating resistance, ultimately paving the way for more effective and personalized therapeutic strategies.
From Theory to Bench: Implementing Modern Overexpression and Knockdown Screens
Traditional genetic screens have long relied on methods that create permanent, binary changes—either completely disrupting a gene's function, as with CRISPR knockout (CRISPRko), or introducing exogenous genetic material for overexpression. While powerful, these approaches have inherent limitations. The advent of CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) has revolutionized functional genomics by enabling reversible, tunable, and precise control over endogenous gene expression without altering the underlying DNA sequence [37]. These technologies use a catalytically deactivated Cas9 (dCas9) that acts as a programmable DNA-binding platform, directing transcriptional effector domains to specific gene promoters to either enhance (CRISPRa) or repress (CRISPRi) transcription [38]. This shift from permanent editing to transient modulation is particularly valuable for studying essential genes, modeling drug actions, understanding complex genetic networks, and conducting sensitive screens in disease-relevant cell types. This guide provides a comprehensive comparison of CRISPRa and CRISPRi, detailing their mechanisms, performance data against alternative technologies, and practical protocols for their implementation in research and drug development.
Fundamental Mechanisms of CRISPRa and CRISPRi
The Core System: dCas9 and Transcriptional Effectors
At the heart of both CRISPRa and CRISPRi is the catalytically dead Cas9 (dCas9). Through point mutations (D10A and H840A in S. pyogenes Cas9), the nuclease activity of Cas9 is abolished, transforming it into a programmable DNA-binding protein that can be precisely targeted to any genomic locus using a guide RNA (gRNA) [38] [37]. The fundamental difference between CRISPRa and CRISPRi lies in the transcriptional effector domains fused to dCas9.
CRISPRi (Interference): For gene repression, dCas9 is typically fused to repressor domains such as the Krüppel-associated box (KRAB). KRAB recruits repressive chromatin-modifying complexes, leading to histone methylation and the formation of heterochromatin, which effectively silences gene transcription [38] [22] [37]. The dCas9-KRAB fusion binds to the transcription start site (TSS), sterically hindering the binding of RNA polymerase and other transcriptional machinery.
CRISPRa (Activation): For gene activation, dCas9 is fused to transcriptional activators. Early systems used simple domains like VP64 (a tetramer of VP16). However, more potent systems have been developed that recruit multiple distinct activator domains simultaneously. Key advanced systems include [38] [22]:
- SAM (Synergistic Activation Mediator): dCas9-VP64 is co-expressed with a modified gRNA containing MS2 RNA aptamers. These aptamers recruit effector proteins (p65 and HSF1 activation domains fused to the MS2 coat protein), creating a powerful multi-component activator complex [38] [22].
- VPR: A tripartite activator directly fused to dCas9, comprising VP64, the p65 subunit of NF-κB, and the Rta transactivator from Epstein-Barr virus [22].
- SunTag: This system uses a dCas9 fused to an array of peptide epitopes (GCN4). These epitopes recruit multiple copies of an antibody single-chain variable fragment (scFv) fused to VP64, thereby amplifying the activation signal [22] [37].
The following diagram illustrates the core components and mechanisms of these systems.
Optimized Guide RNA (gRNA) Design
A critical factor for the success of CRISPRa/i experiments is the design of the gRNA. Unlike CRISPRko, where gRNAs are typically designed to target early exons, CRISPRa and CRISPRi require targeting specific regions relative to the transcription start site (TSS) [38].
- CRISPRi gRNA Design: The optimal window for CRISPRi repression spans from -50 to +300 base pairs (bp) from the TSS, with the most effective gRNAs targeting the first 100 bp downstream of the TSS [38]. This positioning allows dCas9-KRAB to effectively block the assembly of the transcription machinery.
- CRISPRa gRNA Design: For CRISPRa activation, the optimal targeting window is typically -400 to -50 bp upstream of the TSS [38]. This region often contains promoter and enhancer elements where activator complexes can most effectively recruit the transcriptional machinery.
Systematic tiling screens have led to the development of sophisticated algorithms that incorporate chromatin accessibility, nucleosome positioning, and sequence features to predict highly active gRNAs [39]. This has enabled the creation of optimized, genome-wide libraries such as Calabrese (CRISPRa) and Dolcetto (CRISPRi), which achieve high performance with a compact design of 3-10 gRNAs per gene [40].
Performance Comparison with Alternative Technologies
CRISPRi vs. RNAi and CRISPRko
CRISPRi offers several distinct advantages over RNA interference (RNAi) and traditional CRISPR knockout (CRISPRko) for loss-of-function studies.
Table 1: Comparison of Loss-of-Function Technologies
| Feature | CRISPRi | RNAi | CRISPR Knockout (CRISPRko) |
|---|---|---|---|
| Mechanism | dCas9-repressor blocks transcription at the DNA level [38] [37] | siRNA/miRNA mediates mRNA degradation in the cytoplasm [38] [22] | Cas9 nuclease induces double-strand breaks, leading to frameshift mutations [38] |
| Efficiency & Robustness | High; more robust phenotypes in large-scale screens with fewer off-targets [38] [39] | Variable; prone to incomplete knockdown and off-target effects [38] | High for complete gene disruption |
| Reversibility | Reversible and titratable [37] | Reversible | Permanent |
| Toxicity/Genomic Impact | Minimal; no DNA damage, avoids genomic instability [38] [39] | Minimal | High; can induce cytotoxicity and genomic instability from DNA breaks [38] |
| Target Range | Coding & non-coding genes (lncRNAs) [38] [22] | Primarily coding genes; less efficient for nuclear RNA [38] | Coding & non-coding regions (requires large deletions for some) [38] |
| Application in Essential Gene Studies | Ideal; allows partial knockdown without cell death [38] | Suitable | Suboptimal; complete knockout can be lethal [38] |
Key Comparative Insights:
- CRISPRi vs. RNAi: CRISPRi generates more robust and reliable phenotypes in large-scale screens. It outperforms RNAi by avoiding the off-target effects associated with displacing endogenous small RNAs from the RNA-induced silencing complex (RISC) [38]. Furthermore, CRISPRi can efficiently target long non-coding RNAs (lncRNAs), which are often difficult to study with RNAi [22].
- CRISPRi vs. CRISPRko: CRISPRko is a powerful tool for generating complete and permanent loss-of-function. However, the double-strand breaks it introduces can be cytotoxic and confound screens with DNA damage response phenotypes [38] [39]. CRISPRi, by avoiding DNA cleavage, circumvents this issue and is superior for studying essential genes, as cells can often tolerate a partial knockdown but not a complete knockout [38]. A screen in K562 cells demonstrated that the optimized Dolcetto CRISPRi library performed comparably to the Brunello CRISPRko library in detecting essential genes, highlighting its efficacy [40].
CRISPRa vs. ORF Overexpression
For gain-of-function studies, CRISPRa provides a compelling alternative to traditional open reading frame (ORF) overexpression.
Table 2: Comparison of Gain-of-Function Technologies
| Feature | CRISPRa | ORF Overexpression |
|---|---|---|
| Mechanism | Activates endogenous gene from its native genomic context [37] | Introduces exogenous cDNA copy, often driven by a strong viral promoter (e.g., CMV) [38] [22] |
| Expression Level | Physiological or near-physiological; supraphysiological levels are challenging to achieve [38] | Often supraphysiological, non-physiological |
| Splice Variant Expression | Activates the gene's natural splice variants [38] | Typically overexpresses a single, predefined splice variant |
| Genomic Context | Endogenous; preserves native regulatory elements & chromatin environment [41] | Ectopic; subject to positional effects from random integration |
| Library Scalability | Highly scalable; easier and more cost-effective to synthesize genome-wide libraries [38] | More difficult and expensive to synthesize at genome scale |
Key Comparative Insights:
- Physiological Relevance: CRISPRa upregulates genes in their native genomic context, preserving natural splicing and regulatory elements. This often results in more physiologically relevant expression levels compared to the frequently supraphysiological levels driven by strong viral promoters in ORF overexpression [38] [37]. This makes CRISPRa better suited for modeling diseases and processes where correct gene dosage is critical.
- Comprehensive Screening: CRISPRa is more likely to upregulate the most relevant splice variants, which is particularly advantageous when these are unknown [38]. Furthermore, the synthesis of genome-scale CRISPRa libraries is more straightforward and cost-effective than creating comprehensive ORF libraries [38]. In a direct comparison, the Calabrese CRISPRa library identified more vemurafenib resistance genes than an ORF overexpression screen [40].
Quantitative Performance of Optimized Libraries
The development of optimized sgRNA design rules has led to significant performance improvements in CRISPRa/i screens. The table below summarizes key metrics from validated, genome-scale libraries.
Table 3: Performance Metrics of Optimized Genome-wide Libraries
| Library Name | Modality | Key Design Features | Reported Performance |
|---|---|---|---|
| Brunello [40] | CRISPRko | 4 sgRNAs/gene; designed with Rule Set 2 | Outperformed earlier libraries (GeCKO, Avana); high distinction between essential/non-essential genes (dAUC metric) |
| Dolcetto [40] | CRISPRi | Compact design; optimized sgRNA selection | Performed comparably to CRISPRko (Brunello) in detecting essential genes; outperformed earlier CRISPRi libraries |
| Calabrese [40] | CRISPRa | Optimized sgRNA selection for activation | Identified more vemurafenib resistance genes than the SAM library approach |
| hCRISPRi-v2 [39] | CRISPRi | 5 or 10 sgRNAs/gene; incorporates chromatin and sequence features | Detected >90% of essential genes with minimal false positives; lacked non-specific toxicity from DNA breaks |
| hCRISPRa-v2 [39] | CRISPRa | 5 or 10 sgRNAs/gene; optimized for TSS targeting | Identified 60% more genes affecting growth upon overexpression than v1 library |
Experimental Protocols for CRISPRa/i Screening
The following workflow outlines the key steps for conducting a pooled CRISPRa or CRISPRi screen, a common application for identifying genes involved in a biological process of interest.
Detailed Methodologies
Step 1: Generate a Stable Helper Cell Line
- Objective: Create a cell line that stably expresses the dCas9-effector fusion (e.g., dCas9-KRAB for CRISPRi or dCas9-VPR for CRISPRa). This ensures uniform and consistent expression of the CRISPR machinery across the cell population [38] [42].
- Protocol:
- Select a Vector: Use lentiviral vectors optimized for efficient delivery and stable integration, such as those available from commercial suppliers (e.g., Sigma-Aldrich) [38].
- Transduce Cells: Transduce the target cells (e.g., HEK293, K562, or iPSCs) with the dCas9-effector lentivirus. For difficult-to-transfect cells, lentiviral transduction is the method of choice [38].
- Select and Validate: Apply antibiotics (e.g., puromycin, blasticidin) to select for successfully transduced cells. Validate expression of dCas9-effector via Western blot or fluorescence (if tagged with a reporter like BFP) [42]. For inducible systems, functionality must be validated upon addition of the inducer (e.g., doxycycline or trimethoprim) [42] [43].
Step 2: Deliver the sgRNA Library
- Objective: Introduce a pooled sgRNA library into the helper cell line at a low multiplicity of infection (MOI ~0.3) to ensure most cells receive only one sgRNA [40].
- Protocol:
- Library Selection: Choose a genome-scale library (e.g., Calabrese for CRISPRa, Dolcetto for CRISPRi) or a custom, focused library [40].
- Lentiviral Production: Package the sgRNA library into lentiviral particles in HEK293T cells.
- Transduction: Transduce the helper cell line with the sgRNA library virus. Use a sufficient cell number to maintain ~500x coverage of the library to prevent stochastic loss of sgRNAs [40].
- Selection: Apply selection (e.g., puromycin) for 3-7 days to remove untransduced cells. This time point is often harvested as the "T0" baseline.
Step 3: Apply Selection Pressure and Phenotype Readout
- Objective: Culture the cell population under conditions that enrich for or against cells with specific genetic perturbations.
- Protocol:
- Proliferation/Survival Screen: Passage cells for a set duration (e.g., 2-3 weeks), harvesting population samples at multiple time points to track sgRNA dynamics [22].
- Drug Sensitivity Screen: Split the transduced cell population into treated (with drug/toxin) and untreated (vehicle) cohorts. Culture both in parallel for a defined period [22].
- FACS-Based Screen: Use fluorescent reporters or antibodies to sort cells based on a specific phenotype (e.g., surface marker expression, ROS levels) [22] [43].
- Single-Cell RNA-seq Readout: Use CROP-seq or similar methods to capture both the sgRNA identity and the whole transcriptome of single cells [42] [43].
Step 4: Harvest Genomic DNA and Sequence
- Objective: Quantify the relative abundance of each sgRNA in the final population(s) compared to the T0 baseline.
- Protocol:
- Harvest gDNA: Extract genomic DNA from all time points and experimental conditions using standard methods.
- PCR Amplification: Amplify the sgRNA cassette from the genomic DNA using primers that add Illumina sequencing adapters and sample barcodes.
- Next-Generation Sequencing (NGS): Pool the PCR products and sequence on an Illumina platform to obtain millions of reads, allowing for quantitative counting of each sgRNA [40].
Step 5: Bioinformatic Analysis and Hit Identification
- Objective: Identify sgRNAs and genes that are significantly enriched or depleted under the selection pressure.
- Protocol:
- Read Alignment: Map sequencing reads to the reference sgRNA library to generate count tables for each sgRNA in each sample.
- Normalization and Scoring: Use specialized algorithms (e.g., MAGeCK [43], pinAPL) to normalize counts and calculate fold-changes and statistical significance for each sgRNA and gene.
- Hit Calling: Genes with multiple sgRNAs showing consistent and significant depletion (in a loss-of-function screen) or enrichment (in a gain-of-function or resistance screen) are identified as high-confidence hits.
The Scientist's Toolkit: Essential Research Reagents
Successful execution of a CRISPRa/i screen relies on a suite of key reagents and tools.
Table 4: Essential Reagents for CRISPRa/i Screens
| Reagent / Solution | Function | Examples & Considerations |
|---|---|---|
| dCas9-Effector Construct | Core programmable DNA-binding platform fused to activator/repressor domains. | CRISPRi: dCas9-KRAB [38]. CRISPRa: dCas9-VPR, SAM system, SunTag system [22]. |
| Optimized sgRNA Library | Pooled guides targeting genes of interest; determines screen coverage and specificity. | Genome-wide: Calabrese (CRISPRa), Dolcetto (CRISPRi) [40]. Targeted: Custom libraries for specific pathways. Format: Lentiviral plasmid library. |
| Lentiviral Packaging System | Produces viral particles to deliver genetic material into target cells. | Second/third-generation systems (psPAX2, pMD2.G) for safe production of replication-incompetent virus. |
| Cell Line Engineering Tools | Reagents for selecting and validating stable cell lines. | Antibiotics for selection (puromycin, blasticidin); antibodies for validation (Western blot, FACS). |
| Next-Generation Sequencing (NGS) | Enables quantitative tracking of sgRNA abundance in pooled screens. | Illumina platforms; primers with unique barcodes for multiplexing samples [40]. |
| Bioinformatics Software | For analyzing screen data, normalizing counts, and identifying hit genes. | MAGeCK [43], CERES, pinAPL. |
CRISPRa and CRISPRi have firmly established themselves as premier tools for controlled genetic perturbation. Their ability to provide reversible, titratable, and precise modulation of endogenous gene expression offers a more nuanced and physiologically relevant approach compared to traditional knockout and overexpression methods. As demonstrated in diverse applications—from uncovering neuron-specific vulnerabilities in neurodegenerative disease [43] to identifying drivers of chemoresistance in cancer [22]—these technologies are unlocking new dimensions of biology.
The true power of CRISPRa and CRISPRi is often realized when they are used in combination. Parallel loss-of-function and gain-of-function screens can reveal complementary insights, such as identifying both tumor suppressors (which cause phenotypes when overexpressed) and essential oncogenes (which cause phenotypes when knocked down) in the same cell line [38] [22]. Furthermore, their integration with single-cell omics technologies is paving the way for high-resolution mapping of gene regulatory networks and their functional outcomes [42] [43].
As the field advances, future developments will focus on improving the efficiency and specificity of effector domains, expanding the toolset to include epigenetic modifiers, and adapting these platforms for in vivo therapeutic applications. For researchers and drug developers, mastering CRISPRa and CRISPRi is no longer a niche skill but a fundamental requirement for systematically interrogating gene function and accelerating the pace of discovery in the era of precision biology.
The choice between genome-wide and focused library designs represents a critical first step in the planning of functional genetic screens, directly influencing the scope, resolution, and resource requirements of a study. Genome-wide screens aim to interrogate every gene in the genome, offering an unbiased and comprehensive discovery platform. In contrast, focused screens target a predefined subset of genes—such as those encoding cell surface proteins, kinases, or other specific functional classes—to achieve higher depth, statistical power, and practical efficiency within a particular biological context. The decision is not merely a technicality but a strategic consideration that shapes the biological insights one can attain. Furthermore, the choice of perturbation technology—whether loss-of-function (e.g., CRISPR knockout, RNAi) or gain-of-function (e.g., ORF overexpression)—adds another layer of complexity, as each approach can reveal distinct and complementary aspects of gene function [44]. This guide objectively compares the performance of these different library design strategies, supported by experimental data, to inform their optimal application in biological research and drug development.
Comparative Performance of Screening Approaches
Genome-Wide vs. Focused CRISPR Screens
Direct comparative studies demonstrate that focused libraries can significantly enhance the efficiency of identifying relevant hits in specific biological processes. A systematic comparison of a genome-wide CRISPR library and a surfaceome-focused library (targeting 1,344 cell surface proteins) in a rhinovirus (RV) infection model revealed stark differences in performance. The surfaceome screen markedly outperformed the genome-wide screen, with a ~6-fold higher success rate in identifying hit genes crucial for viral infection [45].
Table 1: Quantitative Comparison of Genome-Wide vs. Surfaceome CRISPR Screen
| Screening Metric | Genome-Wide Screen | Surfaceome Screen |
|---|---|---|
| Total Genes Targeted | 18,421 | 1,344 |
| Percentage of Genes with Significant Hits | 0.54% | 3.2% |
| Key Identified Hit | ICAM-1 (known receptor) | ICAM-1 & novel factor OLFML3 |
| Primary Advantage | Unbiased, comprehensive discovery | Higher hit rate and efficiency for a defined protein class |
Both screens successfully identified ICAM-1, the known receptor for major-group rhinoviruses, validating the approach. However, the surfaceome screen also robustly identified olfactomedin-like 3 (OLFML3) as a novel host dependency factor, a finding missed by the genome-wide screen under its standard significance thresholds. This demonstrates that focused designs can powerfully deconvolute complex biological processes by reducing background noise and increasing the detection sensitivity for relevant pathways [45].
Knockdown/Knockout vs. Overexpression Screens
Loss-of-function (LOF) and gain-of-function (GOF) screens represent complementary approaches for dissecting gene function. LOF screens (using CRISPR knockout or RNAi) identify genes whose activity is necessary for a phenotype, revealing essential pathways and vulnerabilities. In contrast, GOF screens (using cDNA overexpression) identify genes that can drive or enhance a phenotype, uncovering potential activators or mechanisms of resistance.
A seminal systematic comparison of CRISPR-Cas9 knockout and shRNA-based knockdown screens in the K562 chronic myeloid leukemia cell line found that while both techniques effectively identified essential genes with high precision (AUC > 0.90), their results showed surprisingly low correlation and were enriched for distinct biological processes [44]. For example, CRISPR-Cas9 screens more strongly identified genes involved in the electron transport chain as essential, whereas shRNA screens more effectively pinpointed subunits of the chaperonin-containing T-complex. This indicates that each technology can access different aspects of biology, potentially due to differences in the timing and completeness of gene perturbation, or technology-specific off-target effects. Combining data from both LOF technologies using a statistical framework (casTLE) improved performance, yielding a more robust set of essential genes [44].
GOF screens are particularly powerful for discovering genes that can confer new functions or resistance to treatments. A genome-scale GOF screen using a library of nearly 12,000 barcoded human open reading frames (ORFs) in primary human T cells identified positive regulators of proliferation, such as the lymphotoxin beta receptor (LTBR) [46]. This approach is invaluable for engineering cell therapies, as it can uncover synthetic drivers that enhance desired effector functions. Dual genome-wide LOF and GOF screens for regulators of cellular resistance to ATR inhibitors (ATRi) further highlight the power of a combined strategy. This approach comprehensively identified genes that, when either knocked out or overexpressed, altered ATRi resistance, uncovering multiple mechanisms including regulation of apoptosis and replication fork stability [47].
Table 2: Comparison of Loss-of-Function and Gain-of-Function Screening Approaches
| Feature | Loss-of-Function (LOF) Screens | Gain-of-Function (GOF) Screens |
|---|---|---|
| Perturbation Type | CRISPR knockout, CRISPRi, RNAi | CRISPR activation (CRISPRa), cDNA ORF overexpression |
| Biological Question | What genes are necessary for a phenotype? | What genes can induce or enhance a phenotype? |
| Typical Library | sgRNAs or shRNAs targeting all genes | sgRNAs for activation or ORF clones |
| Key Applications | Identifying essential genes, drug targets, vulnerability factors | Discovering synthetic drivers, resistance mechanisms, therapeutic genes |
| Complementary Insight | Identifies MED12 as an ATRi resistance factor when knocked out [47] | Identifies LTBR as a driver of T-cell proliferation when overexpressed [46] |
Experimental Protocols for Key Screen Types
Protocol 1: Focused Surfaceome CRISPR Knockout Screen
This protocol is adapted from the screen that identified OLFML3 as a rhinovirus host factor [45].
- Library Design: Construct a focused sgRNA library targeting a curated set of genes encoding cell surface proteins. The library used in the cited study contained 16,975 sgRNAs targeting 1,344 surface proteins, with 12 sgRNAs per gene to ensure robust coverage [45].
- Library Production: Package the sgRNA library into lentiviral particles. Titrate the virus to determine the multiplicity of infection (MOI).
- Cell Line Engineering: Transduce the target cell line (e.g., H1-Hela cells for rhinovirus infection) with the lentiviral library at a low MOI (e.g., 0.3) to ensure most cells receive a single sgRNA. Select transduced cells with puromycin for 7-10 days.
- Phenotypic Selection:
- Split the library-containing cells into experimental and control groups.
- Challenge the experimental group with the selective agent (e.g., rhinovirus RV-B14 at an MOI of 1.0).
- Maintain the control group under mock conditions.
- Culture cells for the duration of the selection period (e.g., 48 hours for RV-induced cell death).
- Sample Collection and Sequencing: Harvest genomic DNA from the surviving cells in the experimental group and from the control group. PCR-amplify the integrated sgRNA sequences and subject them to next-generation sequencing (NGS).
- Bioinformatic Analysis: Use specialized algorithms (e.g., MAGeCK) to compare the abundance of each sgRNA in the experimental group versus the control. Significantly enriched sgRNAs indicate genes whose knockout confers a survival advantage, pointing to key host factors [45].
Protocol 2: Genome-Wide ORF Overexpression Screen
This protocol is based on the screen that identified LTBR as a synthetic driver of T-cell proliferation [46].
- Library Design: Utilize a genome-wide ORF library, such as one containing ~12,000 full-length human ORFs, each tagged with multiple unique DNA barcodes to track individual clones [46].
- Cell Transduction: Transduce the target cells (e.g., primary human CD4+ and CD8+ T cells from multiple donors) with the ORF library at a low MOI. Use a cell number that ensures high coverage of the library (e.g., 200-fold coverage).
- Phenotypic Selection:
- Stimulate the transduced T cells via the T-cell receptor (e.g., using anti-CD3/CD28 antibodies).
- Culture the cells for a defined period (e.g., 14 days) to allow for proliferation.
- Use fluorescence-activated cell sorting (FACS) to isolate the top fraction of cells exhibiting the highest proliferation (e.g., based on dye dilution) or activation (e.g., based on surface marker expression).
- Barcode Sequencing and Analysis: Extract genomic DNA from the sorted population and the unsorted control population. Amplify and sequence the ORF-associated DNA barcodes. Identify significantly enriched barcodes (and thus ORFs) in the sorted population compared to the control using statistical methods. Enriched ORFs represent genes that enhance T-cell proliferation upon overexpression [46].
Visualization of Screening Strategies and Workflows
The following diagrams illustrate the core workflows and conceptual relationships between the different screening strategies discussed in this guide.
The Scientist's Toolkit: Key Research Reagent Solutions
The execution of high-quality genetic screens relies on a suite of well-validated reagents and tools. The following table details essential materials used in the featured studies.
Table 3: Key Research Reagent Solutions for Genetic Screens
| Reagent / Solution | Function in Screening | Specific Examples & Notes |
|---|---|---|
| CRISPR Knockout Library | Enables systematic gene disruption via Cas9-induced double-strand breaks. | GeCKO v2 library [48]; Brunello library [47]. Designed with 4-12 sgRNAs/gene to ensure robustness. |
| ORF Overexpression Library | Enables systematic gene activation through expression of full-length cDNA. | Lentiviral library of ~12,000 barcoded human ORFs [46]. Allows tracking of clones via unique barcodes. |
| Lentiviral Delivery System | Efficiently delivers genetic perturbation elements (sgRNAs, ORFs) into a wide variety of cell types, including primary and non-dividing cells. | Used in nearly all cited studies [45] [46] [48]. |
| Next-Generation Sequencing (NGS) | Quantifies the relative abundance of sgRNAs or ORF barcodes before and after selection to identify hits. | Illumina sequencing is standard. Essential for deconvoluting pooled screen results [45] [46]. |
| Bioinformatic Analysis Tools | Statistically analyzes NGS data to identify significantly enriched or depleted perturbations. | MAGeCK [45] [48]; casTLE (for combining screen types) [44]; RSA algorithm [47]. |
The strategic selection of a library design—genome-wide for unbiased discovery or focused for deep, efficient interrogation of specific pathways—is paramount to the success of a functional genetic screen. As the experimental data demonstrates, a focused surfaceome screen can achieve a higher hit rate for a targeted process like viral infection, while a genome-wide approach remains the gold standard for discovering entirely novel biology [45]. Furthermore, the choice between LOF and GOF perturbations is not a matter of which is superior, but of which is appropriate for the biological question at hand. These approaches are fundamentally complementary, as evidenced by their low correlation and ability to uncover distinct gene sets and biological processes [44]. The most powerful insights often emerge from integrating multiple screening modalities, such as parallel LOF and GOF screens, which together can provide a systems-level view of genetic regulation and identify complex mechanisms, such as those governing resistance to targeted therapies [47]. By understanding the strengths, limitations, and applications of each strategy, researchers can design more effective screens to accelerate the discovery of novel biological mechanisms and therapeutic targets.
In modern biomedical research, the selection of an appropriate biological model system is paramount for accurately deciphering disease mechanisms and identifying therapeutic targets. Within the context of functional genomics, overexpression and knockdown screens represent powerful, complementary approaches for establishing causal relationships between genes and phenotypes. Overexpression screens identify genes that, when highly expressed, drive disease-relevant processes like tumor growth or therapy resistance. Conversely, knockdown (or knockout) screens reveal genes essential for cell survival, proliferation, or disease maintenance. The choice of model system—whether cancer cell lines, induced pluripotent stem cell (iPSC)-derived neurons, or microglial cultures—significantly influences the translatability of these findings, as each system offers unique advantages and limitations in recapitulating human physiology and disease pathology. This guide provides an objective comparison of these model systems, focusing on their applications in perturbation screens, supported by experimental data and methodological protocols.
Cancer Cell Lines: High-Throughput Screening Powerhouses
Cancer cell lines are in vitro model systems widely used in basic cancer research and drug discovery due to their ability to provide an indefinite source of biological material for experimental purposes [49]. Under the right conditions and with appropriate controls, properly authenticated cancer cell lines retain most of the genetic properties of their cancer of origin, making them valuable surrogates for studying tumor biology [49].
Key Applications in Perturbation Screens
Cancer cell lines are the workhorse models for large-scale functional genomic screens, as demonstrated by several key studies:
- Identifying Oncogenic Drivers via Overexpression: A genome-wide screen in yeast identified 245 genes that cause chromosome instability (CIN) when overexpressed, a phenotype known as dosage CIN (dCIN) [50]. This catalog included human orthologs frequently amplified in tumors. Subsequent validation showed that overexpressing two of these genes, TDP1 (a DNA repair enzyme) and TAF12 (a transcription factor), was sufficient to induce CIN in human cells, confirming their potential role as oncogenic drivers [50].
- Uncovering Resistance Mechanisms via CRISPR Knockout/Activation: Dual genome-wide CRISPR knockout and CRISPR activation screens were employed to identify genes that regulate resistance to ATR inhibitors (ATRi), emerging cancer therapeutics [13]. The knockout screen identified genes whose loss confers resistance, while the activation screen pinpointed genes whose gain-of-function promotes survival. This comprehensive approach revealed diverse resistance mechanisms, including restoration of replication fork progression and prevention of apoptosis [13].
- Linking Genetic Alterations to Drug Response: Large-scale panels like the NCI60, the Cancer Cell Line Encyclopedia (CCLE), and the GlaxoSmithKline (GSK) panel have been extensively characterized with multi-omics data (genomics, transcriptomics) [51]. This allows researchers to correlate baseline genetic alterations—such as mutations, copy number variations, and gene expression levels—with responses to thousands of chemical compounds screened against these cell lines, enabling the prediction of drug sensitivity and resistance [51].
Experimental Data and Comparison
The table below summarizes quantitative findings from key cancer cell line studies employing overexpression or knockdown approaches.
Table 1: Experimental Findings from Perturbation Screens in Cancer Cell Lines
| Target Gene | Model System | Perturbation Type | Key Phenotypic Outcome | Measured Effect | Citation |
|---|---|---|---|---|---|
| P2X7 Receptor | LLC & LA795 (Non-small cell lung cancer cells) | Overexpression | Promoted tumor growth, migration, and invasion | Increased migration and invasion in vitro; enhanced tumor growth in vivo | [27] |
| P2X7 Receptor | LLC & LA795 (Non-small cell lung cancer cells) | Knockdown | Suppressed tumor growth, migration, and invasion; promoted apoptosis | Reduced proliferation and migration in vitro; inhibited tumor growth in vivo | [27] |
| ADAM28 | LNCaP & DU145 (Prostate cancer cells) | Overexpression | Increased proliferation and migration | Significant increase in cell proliferation and migration assays | [24] |
| ADAM28 | LNCaP & DU145 (Prostate cancer cells) | siRNA Knockdown | Decreased proliferation and migration | Significant reduction in cell proliferation and migration assays | [24] |
| TDP1 | HT1080 (Fibrosarcoma cells); Rhabdomyosarcoma lines | Overexpression | Induced Chromosome Instability (CIN) | Increased CIN measurable by cytogenetic methods; siRNA knockdown partially rescued phenotype | [50] |
Detailed Methodologies
Typical Workflow for a CRISPR Activation Screen (as in [13]):
- Library Transduction: A pooled lentiviral library containing guide RNAs (gRNAs) designed to activate ~19,000 human genes is transduced into a cancer cell line (e.g., HeLa) at a low multiplicity of infection to ensure one viral integration per cell.
- Selection and Treatment: Cells are selected with an antibiotic (e.g., puromycin) to generate a stable population. This population is then split and treated with either an ATR inhibitor (VE822 or AZD6738) or a vehicle control (DMSO).
- Outgrowth and Analysis: Cells are cultured for several population doublings under selective pressure. Genomic DNA is extracted from the surviving cell populations.
- Next-Generation Sequencing (NGS): The gRNA sequences are PCR-amplified and quantified via NGS.
- Bioinformatic Analysis: The abundance of each gRNA in the drug-treated group is compared to the control group using algorithms like REDundant siRNA Activity (RSA). gRNAs significantly enriched in the treated group identify genes that, when activated, confer resistance to the drug.
Typical Workflow for a Knockdown/Knockout Screen follows a similar process but uses libraries designed to inactivate genes, with enriched gRNAs in the treated group pointing to genes whose loss promotes survival (resistance), while depleted gRNAs indicate genes whose loss causes sensitivity [13].
Diagram 1: CRISPR screen workflow for identifying drug resistance genes.
Research Reagent Solutions
Table 2: Key Reagents for Cancer Cell Line Perturbation Studies
| Reagent / Tool | Function | Example Use Case |
|---|---|---|
| CRISPR Knockout Library (e.g., Brunello) | Targets 19,114 genes with 4 gRNAs/gene for loss-of-function screens. | Identifying genes whose knockout confers resistance or sensitivity to a drug [13]. |
| CRISPR Activation Library (e.g., Calabrese) | Targets gene promoters with activating gRNAs for gain-of-function screens. | Identifying genes whose overexpression drives drug resistance [13]. |
| Validated siRNA Pools | Transient, sequence-specific mRNA knockdown for candidate gene validation. | Confirming phenotypic effects of gene loss in prostate cancer cells [24]. |
| Expression Plasmids (e.g., pCMV-Tag) | Plasmid vectors for constitutive or inducible cDNA overexpression. | Overexpressing P2X7 or ADAM28 to study oncogenic effects [27] [24]. |
| NCI60 / CCLE Panels | Curated collections of well-characterized cancer cell lines with omics data. | Correlating baseline genetic profiles with drug response data [51]. |
iPSC-Derived Neurons: Human-Relevant Neurobiology Models
iPSC-derived neurons offer a powerful tool to study cellular and molecular mechanisms of human neurological disorders in a patient-specific genomic context [52]. They provide an unlimited source of human neurons, bridging the gap when animal models do not fully recapitulate human disease pathology [53].
Key Applications in Disease Modeling
iPSC-derived neurons are particularly valuable for studying inherited disorders, where patient-specific genetic mutations can be linked to functional changes in the affected cell type.
- Modeling Inherited Pain Syndromes: Sensory neurons derived from patients with Inherited Erythromelalgia (IEM) and Small Fiber Neuropathy (SFN)—caused by gain-of-function mutations in the SCN9A (Nav1.7) and SCN10A (Nav1.8) sodium channel genes—display hyperexcitability and reduced rheobase compared to control neurons [53]. This "disease-in-a-dish" phenotype allows for direct functional assessment of patient-specific pathophysiology.
- Protocol-Dependent Functional Maturation: The differentiation protocol significantly impacts the functional properties of the resulting neurons. For example, the widely used "Chambers" protocol and the commercially available "Anatomic" protocol yield sensory neurons with different electrophysiological characteristics; the Chambers protocol produces predominantly tonic-firing neurons, while the Anatomic protocol generates a different firing pattern [53]. This highlights the need to carefully select a differentiation method that yields neurons with the desired properties for a specific screen or assay.
- Therapeutic Screening and Validation: Patient-derived neurons serve as a platform for drug discovery. For instance, the cellular hyperexcitability in IEM-derived sensory neurons was shown to be reversed by selective Nav1.7 blockade, validating the model for target-based drug screening [53].
Experimental Data and Comparison
Table 3: Experimental Findings from Studies Using iPSC-Derived Neurons
| Disease Model | iPSC Source / Genetic Alteration | Key Phenotypic Outcome | Functional Assay | Citation |
|---|---|---|---|---|
| Inherited Erythromelalgia (IEM) | Patient-derived (SCN9A p.Q875E mutation) | Neuronal hyperexcitability, reduced rheobase | Manual patch clamp | [53] |
| Small Fiber Neuropathy (SFN) | Patient-derived (SCN10A mutation) | Neuronal hyperexcitability, reduced rheobase | Manual patch clamp | [53] |
| Control Sensory Neurons | Healthy subject | Standard firing patterns, higher rheobase | Manual patch clamp, Automated patch clamp | [53] |
| Protocol Comparison | Multiple healthy and patient lines | Chambers protocol: Tonic firing; Anatomic: Different firing patterns | Manual patch clamp | [53] |
Detailed Methodologies
Typical Workflow for Generating and Assessing iPSC-Derived Sensory Neurons (as in [53]):
- Reprogramming: Somatic cells (e.g., fibroblasts or peripheral blood mononuclear cells) from patients or controls are reprogrammed into iPSCs using methods like Sendai virus-mediated delivery of the Yamanaka factors (OCT4, SOX2, KLF4, c-MYC).
- Pluripotency Validation: iPSC clones are rigorously characterized through immunocytochemistry for pluripotency markers (SOX2, OCT4, NANOG), demonstration of differentiation into all three germ layers, and karyotyping to ensure genomic integrity.
- Neural Differentiation: iPSCs are differentiated into sensory neurons using defined protocols. The "Chambers" protocol uses small molecules to directly pattern neural precursors, while the "Anatomic" protocol first induces a naive ectodermal intermediate [53].
- Neuronal Characterization: Differentiated neurons are validated by immunostaining for neuronal markers (e.g., β-III-tubulin, peripherin) and sensory neuron-specific markers (e.g., TRPV1, Nav1.7).
- Functional Assessment: Neuronal excitability is primarily assessed using manual patch clamp to measure action potential firing, rheobase (minimum current to elicit an action potential), and ion channel properties. Automated patch clamp, calcium imaging, and multielectrode arrays (MEA) are used for higher-throughput functional analysis.
Diagram 2: Workflow for disease modeling using iPSC-derived neurons.
Microglia: Modeling CNS Immunity and Neuroinflammation
Microglia, the resident immune cells of the central nervous system, play vital roles in brain development, homeostasis, and neurodegenerative diseases. Their complex ontogeny and high degree of heterogeneity make modeling them particularly challenging [54] [55].
Key Model Systems and Applications
Multiple in vitro models have been developed to study microglial biology, each with distinct advantages for functional studies.
- Immortalized Microglial Cell Lines (e.g., HMC3, BV-2): These lines are generated by SV40-dependent immortalization of human or mouse primary microglial cultures [55]. They express many microglial markers (IBA1, CD68, CD11b) and respond to inflammatory stimuli. Their primary advantage is ease of culture, high uniformity, and suitability for high-throughput knockdown/overexpression screens. However, they are a poor representation of ramified, homeostatic microglia as they resemble an activated state and have altered characteristics due to immortalization [55].
- Primary Microglial Cells: Isolated from rodent (typically postnatal) or human brain tissue, primary microglia are considered the gold standard for in vitro studies as they most closely resemble in vivo cells. They are used for detailed functional assays, including phagocytosis, cytokine release, and electrophysiology. However, their use in large-scale screens is limited by difficult isolation, finite lifespan, and phenotypic changes in culture, where they lose many native functions and become more like peripheral macrophages [55].
- Induced Microglia-like Cells (iMGLs): This state-of-the-art model involves differentiating human iPSCs first into hematopoietic progenitor cells and then into microglia-like cells, recapitulating their true yolk-sac ontogeny [55]. iMGLs closely mimic the transcriptional and functional profile of human adult microglia, including ramified morphology, dynamic motility, and appropriate cytokine responses. They represent a powerful, scalable model for studying human-specific microglial biology in a patient-specific context, making them ideal for CRISPR-based screens in neurodegenerative disease.
Research Reagent Solutions
Table 4: Key Model Systems and Tools for Microglial Research
| Model / Tool | Key Characteristics | Best Suited For |
|---|---|---|
| Immortalized Cell Lines (HMC3, BV-2) | Easy culture, high yield, uniform population, express key markers. | High-throughput drug/cytokine screens; initial, rapid knockdown/overexpression studies. |
| Primary Microglial Cultures | Closest in vitro representation of in vivo microglia (from rodents). | Detailed mechanistic studies on phagocytosis, signaling, and physiology. |
| Induced Microglia-like Cells (iMGL) | Human origin, patient-specific, recapitulate ontogeny and transcriptional profile. | Modeling human-specific disease mechanisms; future application in genetic screens. |
| Human-Mouse Chimeric Models | Human iMGLs engrafted into mouse brain. | Studying human microglial function in a complex in vivo environment. |
The choice of a model system is a strategic decision that directly impacts the scope, scale, and biological relevance of findings from overexpression and knockdown screens.
- Cancer Cell Lines are the undisputed champions of scale and throughput. Their ability to be cultured indefinitely in large quantities makes them ideal for genome-wide CRISPR screens. The pre-existing, deeply characterized panels like CCLE allow for immediate correlation of genetic background with phenotypic outcomes. However, they often lack the cellular complexity and native microenvironment of tumors.
- iPSC-Derived Neurons provide an unparalleled window into human-specific neurobiology and genetics. They are indispensable for studying the functional consequences of patient-derived mutations in the relevant cell type, moving beyond overexpression artifacts to observe endogenous pathophysiology. The main challenges include protocol variability, maturation time, and the complexity of recapitulating full neuronal diversity in a dish.
- Microglial Models span a spectrum from simple, high-throughput lines to complex, physiologically relevant iMGLs. The field is rapidly moving towards iPSC-derived models to capture human-specific innate immune responses in the brain, though immortalized lines remain useful for initial, large-scale perturbation experiments.
In conclusion, the integration of data from all three model systems—each with its unique strengths—will provide the most robust and translatable insights. Cancer cell lines offer scalability, iPSC-derived neurons offer human-genetic fidelity, and advancing microglial models are unlocking the complexities of brain immunity. The continued refinement of these tools, especially in improving the maturation and complexity of iPSC-derived models, will further empower researchers to deconvolute disease mechanisms and identify novel therapeutic targets.
The ataxia telangiectasia and Rad3-related (ATR) kinase is a master regulator of the cellular response to DNA damage and replication stress [47] [13]. Cancer cells, due to their high proliferation rates and frequent oncogene activation, experience heightened levels of replication stress, making them particularly dependent on the ATR pathway for survival [47] [56]. This dependency creates a therapeutic window, making ATR an attractive anticancer target [57]. Several ATR inhibitors (ATRis), such as AZD6738 (Ceralasertib) and VE-822 (Berzosertib), have advanced into clinical trials, showing promise both as single agents and in combination with other DNA-damaging therapies [47] [13] [56].
However, as with many targeted therapies, the emergence of resistance poses a significant clinical challenge. A comprehensive understanding of the genetic determinants that drive resistance is paramount for developing strategies to overcome it and for identifying patient populations most likely to respond. This case study examines how a powerful functional genomics approach—dual genome-wide CRISPR knockout (CRISPRko) and CRISPR activation (CRISPRa) screening—was employed to systematically identify genes that regulate cellular resistance to ATR inhibitors [47] [13]. This dual screening paradigm provides a unique lens through which to compare the complementary insights gained from loss-of-function (knockout) and gain-of-function (overexpression) screens within a single, cohesive research framework.
Experimental Design and Workflow
The study was designed to move beyond previous screens that primarily identified genes whose loss caused sensitivity to ATRi. Instead, it focused on uncovering mechanisms of resistance by employing a dual screening strategy in both transformed (HeLa) and non-transformed (MCF10A) cell lines, using two distinct ATR inhibitors (VE822 and AZD6738) to distinguish compound-specific effects from general mechanisms [47] [13].
Screening Libraries and Core Methodology
The experimental workflow relied on two well-established, genome-scale CRISPR libraries.
- CRISPRko Screen: The Brunello human CRISPR knockout library was used to target 19,114 genes with 76,441 unique guide RNAs (gRNAs) [47]. This screen aimed to identify genes whose inactivation confers resistance, suggesting that the normal function of these genes promotes ATRi-induced cell death.
- CRISPRa Screen: The Calabrese human CRISPR activation library was used to overexpress genes by recruiting transcriptional activators to their promoters [47]. This screen aimed to identify genes that, when overexpressed, allow cells to survive ATRi treatment.
A critical design feature was the use of a high dose of ATRi (killing ~90% of cells) to intensely select for resistant clones and powerfully reveal resistance mechanisms [47]. The following diagram illustrates the integrated workflow of this dual screening approach.
Key Research Reagents and Solutions
The following table details the core reagents and tools that formed the backbone of this experimental setup, providing a resource for researchers seeking to implement similar screens.
Table 1: Essential Research Reagents for Dual CRISPR ATRi Resistance Screens
| Reagent / Solution | Function in the Experiment | Specific Examples / Details |
|---|---|---|
| CRISPRko Library | To induce loss-of-function mutations and identify genes whose knockout confers resistance. | Brunello library (targets 19,114 genes with 76,441 gRNAs) [47]. |
| CRISPRa Library | To induce gene overexpression and identify genes whose activation confers resistance. | Calabrese library [47]. |
| ATR Inhibitors | To apply selective pressure and isolate resistant cell populations. | VE822 (Berzosertib) and AZD6738 (Ceralasertib) [47] [13]. |
| Cell Lines | To model both cancerous and non-transformed cellular contexts. | HeLa (cervical cancer) and MCF10A (breast epithelial) [47] [13]. |
| Bioinformatics Algorithm | To statistically analyze sequencing data and rank significant genes. | Redundant siRNA Activity (RSA) algorithm [47]. |
Key Findings and Mechanistic Insights
The dual-screen approach yielded a robust set of candidate genes and revealed diverse biological processes involved in ATRi resistance.
Validation of Knockout Screen Hits
The CRISPRko screen identified numerous genes whose loss promoted survival under ATRi pressure. A significant overlap of 118 genes was found between the top hits for VE822 and AZD6738, indicating common resistance mechanisms [47]. Seven genes (KNTC1, EEF1B2, LUC7L3, SOD2, MED12, RETSAT, and LIAS) were common within the top 40 hits for both inhibitors [47].
Table 2: Top Validated Resistance Genes from the CRISPRko Screen
| Gene | Validated Effect on ATRi Resistance | Reported/Putative Function |
|---|---|---|
| MED12 | Knockdown increased colony formation and cell survival post-ATRi [47]. | Transcriptional regulator; implicated in TGFβ signaling and replication fork stability [47]. |
| KNTC1 | Knockdown increased colony formation and cell survival post-ATRi [47]. | Component of the kinetochore, involved in mitotic chromosome segregation. |
| EEF1B2 | Knockdown increased colony formation and cell survival post-ATRi [47]. | Protein translation elongation factor. |
| SOD2 | Knockdown increased colony formation and cell survival post-ATRi [47]. | Mitochondrial superoxide dismutase, protects against oxidative stress. |
| LUC7L3 | Knockdown increased colony formation and cell survival post-ATRi [47]. | RNA splicing factor. |
| RETSAT | Knockdown increased colony formation and cell survival post-ATRi [47]. | Retinol saturase, involved in vitamin A metabolism. |
| LIAS | Knockdown increased colony formation and cell survival post-ATRi [47]. | Lipoic acid synthetase, involved in mitochondrial metabolism. |
Functional validation via siRNA knockdown confirmed that loss of any of these seven genes significantly increased resistance to both ATR inhibitors, as measured by clonogenic assays and cell proliferation assays (e.g., CellTiter-Glo) [47]. Pathway analysis of the top hits implicated key biological processes, including protein translation, DNA replication, and sister chromatid cohesion [47].
Elucidating the MED12-TGFβ Resistance Mechanism
A particularly insightful finding was the role of MED12, a subunit of the Mediator complex. Further investigation revealed that MED12 knockdown inhibits the TGFβ signaling pathway, which in turn helps to stabilize DNA replication forks and prevent apoptosis during ATR inhibition [47]. This discovery highlights how genetic perturbation can uncover non-obvious signaling networks that drive drug resistance. The following diagram summarizes this mechanistic insight.
Comparative Analysis: Knockout vs. Activation Screens
This dual-screen design allows for a direct comparison of the two approaches, highlighting their synergistic value in a comprehensive research program.
Table 3: Comparison of CRISPRko and CRISPRa Screens in Identifying ATRi Resistance Mechanisms
| Aspect | CRISPR Knockout (CRISPRko) Screen | CRISPR Activation (CRISPRa) Screen |
|---|---|---|
| Primary Goal | Identify genes whose loss-of-function confers resistance. | Identify genes whose gain-of-function confers resistance. |
| Biological Insight | Reveals genes that are necessary for ATRi efficacy in their normal state (e.g., pro-apoptotic genes). | Reveals genes that can bypass the need for ATR or activate compensatory survival pathways. |
| Therapeutic Implication | Inactivation of these genes in tumors may predict innate or acquired resistance. | Overexpression of these genes in tumors may serve as a biomarker for resistance. |
| Mechanisms Identified | Restoration of replication fork progression, prevention of apoptosis (e.g., via MED12-TGFβ) [47]. | Potential activation of parallel DNA damage response (DDR) pathways or pro-survival signals. |
| Complementarity | Answers: "What breaks in the cell's machinery allows it to survive ATRi?" | Answers: "What new tools can the cell use to overcome ATRi?" |
The data from this study demonstrates that knockout and activation screens are not redundant; they interrogate the genome from two opposing and complementary angles. While the knockout screen identified genes like MED12 that normally sensitize cells to ATRi, the activation screen (though less detailed in the available results) would point to genes that can actively drive resistance when upregulated. This combined data provides a more complete picture of the potential resistance landscape in a single experiment.
Broader Research Context and Validation
The utility of CRISPR screens for understanding ATRi response is reinforced by independent studies. For instance, a genome-wide CRISPRko screen in soft-tissue sarcoma cells identified genes within the ATM signaling network as critical determinants of sensitivity to AZD6738 [57]. This independent finding confirmed the close interplay between the ATR and ATM pathways and suggested that ATM inhibition could synergize with ATRi to overcome resistance [57], a strategy supported by in vivo models.
From a methodological standpoint, the choice of analysis software is critical for robust hit-calling in CRISPR screens. The field has developed numerous algorithms, with MAGeCK and RSA (used in the featured study) being among the most common [58] [59]. Benchmarking studies suggest that while different tools have varying strengths, methods based on negative binomial models (like MAGeCK) often demonstrate strong performance [59].
This case study exemplifies the power of dual CRISPRko/CRISPRa screening as a comprehensive strategy for dissecting complex drug resistance mechanisms. By simultaneously probing both loss and gain of function, the study:
- Identified a high-confidence set of genes, including MED12, that drive ATR inhibitor resistance.
- Uncovered novel biology, linking the MED12-TGFβ axis to replication fork stability and cell survival under replication stress.
- Provided a rich dataset of potential biomarkers that could guide the clinical development of ATR inhibitors toward patients most likely to benefit.
The comparative framework of knockout versus activation screens proves to be a superior approach, offering a more holistic view of the genetic networks that control a cell's fate upon therapeutic challenge than either method could provide alone. This paradigm is readily applicable to the study of other targeted therapies, accelerating the journey toward personalized cancer medicine.
Microglia, the resident immune cells of the central nervous system, are emerging as key drivers of neurological diseases, yet a systematic understanding of their underlying mechanisms has remained challenging [42]. The development of advanced CRISPR-based functional genomics has created new opportunities to bridge the gap between disease-associated genetic variants and changes in microglial function [60]. This case study examines a specific CRISPR interference/activation (CRISPRi/a) platform in human induced pluripotent stem cell (iPSC)-derived microglia, highlighting how this technology enables systematic discovery of microglial state regulators. The platform exemplifies the distinct and complementary insights that can be gained from knockdown (CRISPRi) and overexpression (CRISPRa) screens, offering a powerful comparative approach for functional genomics research [42].
Platform and Protocol Design
Engineered iPSC Line and Differentiation Strategy
To overcome challenges inherent in lengthy microglia differentiation protocols that create population bottlenecks unsuitable for pooled screens, researchers developed an innovative transcription factor-driven approach [42].
Key Engineering Steps:
- An iPSC line was engineered with two integrated cassettes for doxycycline-inducible expression of six microglia-associated transcription factors: PU.1, MAFB, CEBPα, CEBPβ, IRF5, and IRF8 [42].
- These factors were integrated into the CLYBL and AAVS1 safe-harbor loci to ensure stable expression [42].
- The platform incorporated inducible CRISPRi or CRISPRa machinery into the same iPSC line, enabling genetic perturbations after differentiation [42].
Differentiation Protocol: The established three-step protocol generates microglia-like cells (iTF-Microglia) in only 8 days [42]:
- Day 0: Doxycycline induction of transcription factor expression begins
- Day 2: Medium supplementation with GM-CSF and IL-34
- Day 4: Additional supplementation with M-CSF and TGF-β
- Day 8: Fully ramified microglia morphology achieved with excellent viability maintained for at least 8 more days [42]
CRISPRi/a System Specifications
The platform features three distinct configurations for flexible experimental design [42]:
Table: CRISPRi/a System Configurations
| System Type | Core Components | Control Mechanism | Primary Applications |
|---|---|---|---|
| Constitutive CRISPRi | CAG promoter-driven dCas9-BFP-KRAB | Always active | Continuous gene knockdown |
| Inducible CRISPRi | DHFR degron-tagged dCas9-KRAB | TMP-stabilized | Temporally controlled knockdown |
| Inducible CRISPRa | DHFR-dCas9-VPH | TMP-stabilized | Temporally controlled gene activation |
Platform Engineering and Differentiation Workflow
Experimental Methodologies and Screening Approaches
Functional Characterization of iTF-Microglia
The iTF-Microglia were rigorously validated to ensure they recapitulate essential microglial functions [42]:
Phagocytosis Assay:
- Cells were incubated with fluorescent beads or rat synaptosomes
- Phagocytosis was quantified by measuring fluorescence intensity
- Specificity was confirmed using actin polymerization inhibitor Cytochalasin D
- Results demonstrated robust, actin-dependent phagocytic capability [42]
Inflammatory Response Profiling:
- LPS stimulation for 24 hours induced characteristic morphological changes from ramified to ameboid
- RNA sequencing identified upregulation of immune response genes (C3, CXCL10, IL32, SAA1)
- Cytokine array measuring 36 secreted factors showed elevated CCL2 and CXCL1 under control conditions, and significant increases in IL-6 (14-fold), IL-8, and CXCL10 following LPS stimulation [42]
Neuronal Coculture System:
- iTF-Microglia were cocultured with iPSC-derived glutamatergic neurons
- Cocultured microglia maintained pronounced ramified morphology, demonstrating physiological relevance [42]
Pooled Screening Methodologies
The platform enabled three distinct screens targeting the "druggable genome" [42]:
Survival/ Proliferation Screen:
- sgRNA library introduced into iTF-Microglia
- Cells collected at multiple time points
- sgRNA abundance changes quantified by sequencing to identify genes essential for microglial survival [42]
Phagocytosis Screen:
- Cells incubated with fluorescent synaptosomes or beads
- High- and low-phagocytosis populations isolated by fluorescence-activated cell sorting (FACS)
- Differential sgRNA abundance identified regulators of phagocytic function [42]
Activation Screen:
- LPS stimulation to induce inflammatory activation
- Sorting based on activation markers or morphological changes
- sgRNA sequencing identified modifiers of inflammatory responses [42]
Single-Cell RNA Sequencing Screen:
- Pooled CRISPR perturbations combined with single-cell RNA sequencing readout
- Enabled mapping of transcriptional states resulting from genetic perturbations
- Identified regulators of microglial state transitions [42]
Key Findings and Comparative Data
Screening Results and Hit Validation
The CRISPRi/a screens uncovered numerous regulators of microglia biology, with particular emphasis on genes associated with neurodegeneration [42]. A disease-associated microglial state characterized by osteopontin (SPP1) expression was identified and shown to be selectively depleted by colony-stimulating factor-1 (CSF1R) inhibition, demonstrating the platform's utility for identifying therapeutic targets [42].
Table: Comparative Screening Results for Selected Microglial Processes
| Target Process | CRISPRi Hits (Knockdown) | CRISPRa Hits (Overexpression) | Neurodegeneration Association |
|---|---|---|---|
| Cell Survival | Essential genes identified | Survival-promoting genes | TREM2, APOE-related pathways |
| Phagocytosis | Positive and negative regulators | Enhanced phagocytosis mediators | Alzheimer's disease risk genes |
| Inflammatory Activation | Activation suppressors | Activation drivers | Pro-inflammatory cytokine networks |
| State Transitions | State-stabilizing factors | State-promoting factors | Disease-associated microglia (DAM) genes |
Advantages of Parallel Knockdown and Overexpression Screens
The parallel implementation of CRISPRi and CRISPRa provided complementary insights [42] [60]:
CRISPRi (Knockdown) Strengths:
- Identifies essential genes and pathways required for microglial functions
- Models loss-of-function mutations and haploinsufficiency
- Reveals genes whose reduction may be protective
- Fewer off-target effects compared to RNAi [60]
CRISPRa (Overexpression) Strengths:
- Identifies genes whose enhanced expression drives functional changes
- Models disease-associated gene duplications or enhanced signaling
- Reveals potential therapeutic targets for activation
- Can identify suppressors of pathological states [42]
Synergistic Value:
- Orthogonal validation of screening hits across both platforms
- Distinguishes between essential genes and those with dosage-sensitive effects
- Provides comprehensive view of pathway regulation
- Informs therapeutic strategy (inhibition vs activation) [42] [60]
Comparative Screening Approach and Applications
The Scientist's Toolkit: Essential Research Reagents
Table: Key Research Reagents for CRISPRi/a Microglial Screening
| Reagent / Solution | Function / Application | Specifications / Alternatives |
|---|---|---|
| iPSC Line with Integrated TFs | Microglia differentiation foundation | Six TF (PU.1, MAFB, CEBPα, CEBPβ, IRF5, IRF8) in CLYBL/AAVS1 loci |
| Inducible CRISPRi Machinery | Gene knockdown | DHFR-degron tagged dCas9-KRAB, TMP-controlled |
| Inducible CRISPRa Machinery | Gene activation | DHFR-dCas9-VPH, TMP-controlled |
| Druggable Genome Library | Targeted sgRNA collection | Focused on therapeutically relevant targets |
| Differentiation Cytokines | Microglia maturation | GM-CSF, IL-34, M-CSF, TGF-β supplementation |
| Phagocytosis Substrates | Functional assay | Fluorescent beads, rat synaptosomes |
| Activation Stimuli | Inflammatory challenge | LPS for pro-inflammatory activation |
Discussion and Research Implications
Technical Advantages of the Platform
This CRISPRi/a platform addresses several critical challenges in microglial functional genomics [42]:
Scalability and Efficiency:
- The 8-day differentiation protocol avoids population bottlenecks that skew sgRNA library representation in lengthier protocols
- Enables genome-wide screens in authentic human microglial cells
- Compatible with multiple phenotypic readouts including scRNA-seq [42]
Genetic Tool Versatility:
- Inducible systems enable temporal control of perturbations
- Multiple screening modalities (loss- and gain-of-function) in identical genetic background
- Direct comparison of knockdown vs overexpression effects on same genes [42] [60]
Physiological Relevance:
- Human iPSC-derived system captures human-specific biology
- Functional validation in physiological processes (phagocytosis, inflammation)
- Neuronal coculture capability for microenvironmental context [42]
Comparison with Alternative Approaches
Table: Platform Comparison with Alternative Microglial Research Methods
| Methodology | Genetic Control | Human Relevance | Throughput | Key Limitations |
|---|---|---|---|---|
| CRISPRi/a in iTF-Microglia | Precise (endogenous) | High (human iPSC) | High (pooled) | Requires specialized engineering |
| Primary Microglia | Limited (transduction) | High | Low | Donor variability, limited expansion |
| Immortalized Lines | Moderate | Low | High | Transformed phenotype |
| Animal Models | Good (in vivo) | Moderate | Medium | Species differences |
| RNAi in Microglia | Moderate (off-targets) | Variable | Medium | Off-target effects, incomplete knockdown |
Future Research Directions
The platform establishes a foundation for numerous advanced applications [42] [60]:
Disease Modeling:
- Introduction of disease-associated genetic variants via base editing
- Investigation of patient-specific iPSC backgrounds
- Modeling gene-environment interactions in microglial function [60]
Therapeutic Development:
- High-throughput target validation for neurodegenerative diseases
- Identification of state-specific therapeutic targets
- Testing combination therapies targeting multiple pathways [42]
Advanced Screening Modalities:
- Integration of single-cell multi-omics readouts
- Spatial transcriptomics following perturbations
- High-content imaging-based phenotypic screening [42] [60]
This CRISPRi/a platform represents a significant advancement in microglial functional genomics, enabling systematic discovery of regulators governing microglial states in health and disease. The parallel implementation of knockdown and overexpression screens provides complementary insights that enhance target validation and inform therapeutic development strategies. By enabling large-scale genetic screening in human iPSC-derived microglia with both loss-of-function and gain-of-function approaches, this platform addresses a critical methodological gap in neuroimmunology and neurodegeneration research. The ability to identify not only essential genes but also those whose manipulation can potentially reprogram microglial states offers particular promise for developing novel therapeutic approaches for neurodegenerative diseases where microglial dysfunction plays a central role.
Phenotypic screening has re-emerged as a powerful, unbiased strategy for identifying bioactive compounds and gene functions based on observable effects in cells, tissues, or whole organisms, without requiring prior knowledge of a specific molecular target [61]. In functional genomics, this approach allows researchers to connect genetic perturbations to functional outcomes, providing a systems-level understanding of gene function [62]. The core principle involves subjecting cells or organisms to genetic perturbations—typically knockdown (CRISPRi) or overexpression (CRISPRa)—and then measuring the resulting phenotypic changes using a diverse array of readouts [62] [63]. These readouts range from simple metrics like cell survival to highly multiplexed profiles capturing morphological, transcriptomic, and proteomic changes [64].
The choice between knockdown and overexpression screens depends heavily on the biological question and the phenotypic readouts employed. Knockdown screens (using CRISPRn or CRISPRi) are ideal for identifying essential genes, validating drug targets, or understanding loss-of-function phenotypes [62]. In contrast, overexpression screens (using CRISPRa) can identify genes that, when upregulated, confer a selective advantage or resistance, reveal gene functions masked by redundancy, or discover potential therapeutic genes [62]. The sophistication of phenotypic readouts has advanced dramatically, moving beyond single-parameter measurements to massively multiplexed profiling that can capture the complexity of biological systems [64] [61]. This guide provides a comparative analysis of these readouts and their effective application in both knockdown and overexpression screening paradigms.
Comparative Analysis of Phenotypic Readouts
The selection of an appropriate phenotypic readout is critical for extracting meaningful biological insights from functional genomics screens. Different readouts vary in their complexity, throughput, informational depth, and suitability for knockdown versus overexpression approaches.
Table 1: Comparison of Common Phenotypic Readouts in Functional Genomics Screens
| Phenotypic Readout | Typical Assay Methods | Throughput | Key Applications | Suitability for Knockdown Screens | Suitability for Overexpression Screens |
|---|---|---|---|---|---|
| Cell Survival & Proliferation | Colony formation, ATP-based viability, dye incorporation [62] | High | Identification of essential genes; drug modifier screens [62] | Excellent for identifying essential genes and synthetic lethal interactions [62] | Excellent for identifying genes conferring proliferation advantage or drug resistance [62] |
| Phagocytosis | Uptake of pH-sensitive particles (e.g., pHrodo-labeled synaptosomes), fluorescent bacteria [62] [65] | Medium | Neurobiology, immunology, host-pathogen interactions [62] [65] | Good for identifying negative regulators of phagocytic pathways [62] | Good for identifying positive regulators that enhance phagocytic capacity [62] |
| Transcriptomics | Single-cell RNA sequencing (e.g., Perturb-seq, CROP-seq) [62] | Medium (increasing) | Unbiased pathway mapping; understanding signaling networks [62] | Excellent for mapping gene regulatory networks and downstream effects of loss-of-function [62] | Excellent for identifying transcriptional programs activated by gene overexpression [62] |
| High-Content Morphological & Multiplexed Profiling | High-content imaging, Cell Painting, in-situ sequencing [64] | Medium | Comprehensive phenotypic profiling; drug discovery; pathway analysis [64] [63] | Excellent for detecting subtle morphological changes and complex phenotypes from loss-of-function [64] | Excellent for identifying morphological changes induced by gene activation [64] |
| Cell Surface Marker Expression | Flow Cytometry (FACS), Magnetic-Activated Cell Sorting (MACS) [62] | High | Differentiation status, immune cell profiling, signaling activity [62] | Good for identifying regulators of differentiation and cell identity [62] | Good for inducing or enhancing differentiation markers [62] |
The data from diverse phenotypic readouts is not redundant; rather, it provides complementary information that significantly enhances gene function prediction. Research in yeast models has demonstrated that genetic interaction networks mapped using different phenotypic readouts overlap less than expected and each provides unique functional information [66]. Combining these networks improves the accuracy of gene function prediction, demonstrating that employing multiple distinct phenotypic readouts can substantially increase the biological insights gained from genetic screens [66].
Experimental Protocols for Key Phenotypic Readouts
Cell Survival and Proliferation Assay
Purpose: To identify genes essential for cell viability or that modify sensitivity to compounds and other insults [62].
Workflow:
- Library Transduction: Transduce cells with a genome-wide CRISPR knockout (CRISPRn) or activation (CRISPRa) library at an appropriate MOI to ensure single-guide integration [62].
- Selection: Apply appropriate selection (e.g., puromycin) to eliminate untransduced cells.
- Phenotype Induction: Culture cells for multiple population doublings (typically 10-14 days) to allow depletion or enrichment of guide RNAs affecting viability. For drug screens, add the compound of interest at the desired concentration [62].
- Sample Collection: Harvest cell pellets at the beginning (T0) and end (Tfinal) of the experiment.
- Genomic DNA Extraction & NGS Library Prep: Isolate genomic DNA, amplify the integrated guide RNA sequences, and prepare for next-generation sequencing [62].
- Data Analysis: Sequence the guide RNA inserts and quantify their abundance in T0 versus Tfinal samples. Depleted guides in knockdown screens indicate essential genes, while enriched guides in overexpression screens may indicate genes conferring growth advantage [62].
Phagocytosis Assay in Microglia
Purpose: To identify genetic regulators of phagocytic function, particularly relevant in immune cells and disease models [62] [65].
Workflow:
- Genetic Perturbation: Differentiate human pluripotent stem cells (hPSCs) into microglia-like cells and introduce focused genetic perturbations (e.g., CRISPRi/a against kinase/phosphatase libraries or transcription factors) [62].
- Challenge with Phagocytic Cargo: Incubate cells with pHrodo Red-labeled synaptosomes or amyloid-β peptides. pHrodo fluoresces brightly in the acidic environment of phagolysosomes, specifically identifying particles that have been internalized [62] [65].
- Quantification: Analyze cells using flow cytometry or high-content imaging to measure the fluorescence intensity associated with each cell, which corresponds to phagocytic activity [62].
- Data Analysis: Compare phagocytosis levels between control and genetically perturbed cells. Hits are identified as perturbations that significantly increase or decrease the median fluorescence intensity compared to non-targeting controls [62].
High-Content Morphological and Multiplexed Profiling
Purpose: To capture rich, multidimensional phenotypic data combining morphology, RNA, and protein expression from the same cells [64].
Workflow:
- Optical Pooled Screening (OPS) Setup: Conduct a pooled CRISPR screen in cells plated for imaging [64].
- In-Situ Sequencing (ISS): Perform targeted in-situ sequencing of the genetic perturbations (guide RNAs) directly in the cells to link phenotypes to genotypes [64].
- Multiplexed Staining:
- Morphology: Apply Cell Painting dye cocktail to label multiple cellular compartments (nucleus, cytoplasm, mitochondria, etc.) [64].
- Protein: Use immunofluorescence with antibody panels to detect specific protein markers (e.g., 50-plex panels) [64].
- RNA: Hybridize targeted RNA probe panels to detect hundreds of transcripts [64].
- Automated Imaging: Acquire high-resolution images of all channels using automated microscopy.
- Feature Extraction & Analysis: Use image analysis software to extract thousands of morphological features and quantify RNA and protein signals. Employ machine learning to create phenotypic embeddings and identify distinct phenotypic clusters associated with each genetic perturbation [64].
Diagram 1: Workflow for high-content morphological and multiplexed profiling in optical pooled screening, enabling linked genotype-phenotype analysis at single-cell resolution [64].
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of phenotypic screens requires carefully selected reagents and tools. The following table details key solutions for setting up these experiments.
Table 2: Essential Research Reagents for Phenotypic Screening
| Reagent / Tool | Function | Example Application |
|---|---|---|
| CRISPR Knockout (CRISPRn) Library | Introduces frameshift mutations to disrupt gene function [62] | Genome-wide loss-of-function screens to identify essential genes [62] |
| CRISPR Interference (CRISPRi) Library | Uses dCas9 fused to repressor domains to silence gene transcription [62] | Knockdown screens in DNA damage-sensitive cells like hPSCs [62] |
| CRISPR Activation (CRISPRa) Library | Uses dCas9 fused to activator domains to enhance gene transcription [62] | Gain-of-function screens to identify genes that confer resistance or drive differentiation [62] |
| pHrodo-based Phagocytosis Reporters | pH-sensitive dyes that fluoresce upon phagolysosome internalization [62] | Specific quantification of phagocytic activity in microglia and macrophages [62] |
| Cell Painting Dye Cocktail | A six-dye mixture that labels multiple organelles for morphological profiling [64] | High-content imaging to capture detailed morphological phenotypes [64] |
| In-Situ Sequencing (ISS) Reagents | Enable sequencing of guide RNAs directly in fixed cells [64] | Genotype-phenotype linking in optical pooled screens [64] |
| Multiplexed FISH/Ab Panels | Targeted RNA probes and antibody panels for multiplexed detection [64] | Simultaneous measurement of hundreds of RNA and protein species in single cells [64] |
| Element AVITI24 Platform | Supports simultaneous detection of protein, RNA, and morphological features [64] | Integrated multimodal profiling in optical pooled screens [64] |
Diagram 2: Complementary biological insights from knockdown versus overexpression screening approaches, each accessing distinct areas of phenotypic space [62].
The expanding repertoire of phenotypic readouts, from traditional cell survival to sophisticated multimodal profiling, provides researchers with powerful tools to deconvolve gene function in health and disease. The choice of readout must be aligned with the specific biological question and the type of genetic screen being performed. Knockdown screens excel at identifying essential genes and loss-of-function phenotypes, while overexpression screens are optimal for discovering genes that drive processes when activated. Critically, employing diverse phenotypic measures in parallel yields complementary information that significantly enhances functional prediction [66]. As technologies like optical pooled screening and single-cell multi-omics continue to mature, they will further bridge the gap between genetic perturbation and systems-level phenotypic understanding, accelerating both basic biological discovery and therapeutic development.
Maximizing Screen Efficacy: Troubleshooting Common Pitfalls and Optimization Strategies
CRISPR interference (CRISPRi) has emerged as a powerful tool for programmable gene knockdown in functional genomics. This guide objectively compares the performance of dual-sgRNA libraries against single-sgRNA and alternative CRISPR approaches, presenting experimental data that demonstrate their enhanced efficacy. Within the broader context of comparing overexpression and knockdown screening methodologies, we examine how dual-sgRNA designs address critical limitations in genetic screening, including incomplete penetrance and high false-negative rates. The data summarized herein provide researchers with evidence-based recommendations for selecting optimal CRISPRi strategies for their specific experimental needs, particularly in drug discovery and functional genomics applications.
The landscape of functional genomics has been revolutionized by CRISPR-based screening technologies, which primarily fall into two categories: loss-of-function (knockdown/knockout) and gain-of-function (overexpression) approaches. CRISPR interference (CRISPRi) enables reversible, titratable gene knockdown without introducing DNA double-strand breaks, making it particularly valuable for studying essential genes and non-coding RNAs [67]. Unlike CRISPR nuclease (CRISPRn) that permanently disrupts genes, CRISPRi uses a catalytically dead Cas9 (dCas9) fused to transcriptional repressor domains to epigenetically silence target genes [67].
A fundamental challenge in genetic screening has been balancing library size with knockdown efficacy. Early CRISPRi screens relied on single-sgRNA libraries containing 3-6 guides per gene to minimize false negatives, resulting in large libraries that constrained experimental scalability [67]. The emergence of dual-sgRNA libraries represents a significant innovation, combining ultra-compact design with enhanced knockdown efficiency through synergistic guide action.
Comparative Performance Analysis of CRISPR Libraries
Direct Comparison of Single vs. Dual-sgRNA Libraries
Experimental data from genome-wide growth screens in K562 cells demonstrate the superior performance of dual-sgRNA designs. When targeting essential genes previously identified by the Cancer Dependency Map (DepMap), dual-sgRNA libraries produced significantly stronger growth phenotypes compared to single-sgRNA libraries [67].
Table 1: Performance Comparison of Single vs. Dual-sgRNA Libraries in Essential Gene Screening
| Library Design | Mean Growth Rate (γ) | P-value | Correlation with Published Screens | AUC for Essential Gene Detection |
|---|---|---|---|---|
| Single-sgRNA | -0.20 | Reference | r = 0.82 | >0.98 |
| Dual-sgRNA | -0.26 | 6 × 10⁻¹⁵ | r = 0.83 | >0.98 |
The 30% improvement in growth suppression with dual-sgRNA libraries (γ = -0.26 vs. -0.20) demonstrates substantially enhanced knockdown efficacy while maintaining perfect recall of essential genes (AUC >0.98) [67]. This compact design enables more complex screens in applications where cell numbers or sequencing costs are limiting.
Benchmarking Against Alternative CRISPR Modalities
Recent benchmarking studies have evaluated dual-sgRNA libraries against other screening approaches. A 2025 benchmark comparison of CRISPRn guide-RNA design algorithms revealed that dual-targeting libraries consistently outperformed single-guide libraries in both essentiality and drug-gene interaction screens [14].
Table 2: Broad Comparison of CRISPR Screening Modalities
| Screening Method | Key Features | Advantages | Limitations |
|---|---|---|---|
| Dual-sgRNA CRISPRi | Two sgRNAs per gene targeting the same locus | High knockdown efficacy, reversible, minimal genomic damage | Potential for template switching during lentiviral production |
| Single-sgRNA CRISPRi | One sgRNA per gene | Smaller library size, well-established protocols | Variable knockdown efficiency between guides |
| CRISPRn (Knockout) | Introduces double-strand breaks | Permanent gene disruption | DNA damage toxicity, genomic rearrangements |
| CRISPRa (Activation) | dCas9 fused to transcriptional activators | Gain-of-function studies | Can induce non-physiological expression levels |
| Arrayed CRISPR | Individual perturbations in separate wells | Suitable for complex phenotypes | Resource-intensive, lower throughput |
Notably, dual-sgRNA libraries demonstrated stronger depletion of essential genes and reduced enrichment of non-essential genes in Chronos gene fitness estimates compared to single-targeting guides [14]. However, researchers should note that dual targeting may trigger a heightened DNA damage response in CRISPRn applications, observed as a modest fitness reduction even in non-essential genes [14].
Experimental Protocols and Workflows
Dual-sgRNA Library Construction and Screening
The experimental workflow for dual-sgRNA screening follows established pooled CRISPR screening methodologies with specific modifications for dual-guide cassettes:
Dual-sgRNA Screening Workflow
Library Design and Cloning:
- Select two highly active sgRNAs per gene based on empirical validation or predictive algorithms like VBC scores [14]
- Clone sgRNAs as tandem cassettes into lentiviral transfer vectors containing puromycin resistance markers
- For CRISPRi, utilize optimized effectors such as Zim3-dCas9 that balance strong on-target knockdown with minimal non-specific effects [67]
Virus Production and Cell Transduction:
- Produce lentiviral particles using third-generation packaging systems in HEK293T cells
- Transduce target cells at low multiplicity of infection (MOI = 0.3-0.6) to ensure most cells receive only one sgRNA combination
- Maintain at least 200x library coverage throughout the experiment to preserve library representation [68]
Selection and Analysis:
- Apply puromycin selection 24 hours post-transduction to eliminate untransduced cells
- Harvest cells at multiple time points for phenotypic assessment (e.g., growth rates, drug resistance)
- Extract genomic DNA and amplify sgRNA cassettes using a two-step PCR protocol compatible with next-generation sequencing [67]
- Analyze sgRNA abundance changes using specialized algorithms (MAGeCK, Chronos) to identify hit genes [69]
Advanced Library Designs: From Dual to Quadruple-sgRNA
Recent innovations have further expanded beyond dual-sgRNA designs. The development of quadruple-guide RNA (qgRNA) libraries through Automated Liquid-Phase Assembly (ALPA) cloning demonstrates additional improvements in perturbation efficacy [70]. In gene activation experiments, qgRNA vectors massively increased target gene activation compared to individual sgRNAs, particularly for genes with low basal expression levels [70]. This multi-guide approach achieved 75-99% efficacy in deletion experiments and 76-92% in silencing experiments, representing the current state-of-the-art in CRISPR library design [70].
Mechanisms of Enhanced Efficacy
The superior performance of dual-sgRNA libraries stems from multiple synergistic mechanisms:
Enhanced Knockdown Through Synergistic Repression: In CRISPRi applications, targeting two independent sites within a gene promoter likely recruits higher local concentrations of repressive complexes, leading to more complete transcriptional shutdown [67]. This multi-target approach mitigates the variability in efficacy observed between individual sgRNAs.
Increased Probability of Complete Gene Disruption: For CRISPRn applications, dual-sgRNA designs increase the probability of generating functional knockouts through larger deletions between target sites. When two adjacent genomic sites are cut simultaneously, cellular repair mechanisms may generate deletions spanning both sites, almost guaranteeing frameshift mutations and complete gene disruption [14].
Reduced False Negatives from sgRNA Inefficiency: Individual sgRNAs exhibit variable efficacy due to sequence context, chromatin accessibility, and other factors. By including two independent guides per gene, dual-sgRNA designs ensure that if one guide performs suboptimally, the other may still mediate effective knockdown, substantially reducing false-negative rates in screens [67].
Applications in Drug Discovery and Functional Genomics
Dual-sgRNA libraries have enabled more robust genetic screening across diverse applications:
Identification of Synthetic Lethal Interactions: In cancer research, dual-sgRNA screens have identified genotype-specific dependencies, revealing therapeutic targets for precision oncology. For example, genome-wide screens in PARP10-overexpressing breast cells identified synthetic lethal interactions with DNA repair factors including ATM, suggesting potential combination therapies [69].
Studying Essential Genes and Pathway Analysis: The enhanced knockdown efficacy enables more precise characterization of essential biological pathways. In studies of DNA damage response, dual-sgRNA CRISPRi screens revealed that PARP10 impacts ATM recruitment to nascent DNA upon replication stress, elucidating novel regulatory mechanisms [69].
Complex Model Systems: The compact size of dual-sgRNA libraries facilitates screening in challenging models such as organoids and in vivo systems, where large library complexity presents practical limitations [71] [72]. The reduced library size enables maintenance of sufficient coverage in systems with limited cell numbers.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Dual-sgRNA Screening
| Reagent/Resource | Function | Examples/Specifications |
|---|---|---|
| Zim3-dCas9 | Optimized CRISPRi effector | Provides strong on-target knockdown with minimal non-specific effects on cell growth/transcriptome [67] |
| Dual-sgRNA Lentiviral Libraries | Gene perturbation | Ultra-compact designs (1-3 elements per gene) with high activity; available from Jostlab and Weissman labs [67] |
| ALPA Cloning System | High-throughput plasmid assembly | Enables efficient construction of multi-sgRNA arrays; uses dual antibiotic selection [70] |
| VBC Score Algorithm | sgRNA efficacy prediction | Computational tool for selecting highly active guides; correlates with experimental efficacy [14] |
| MAGeCK & Chronos | Bioinformatic analysis | Algorithms for identifying significantly enriched/depleted sgRNAs from screen data [69] [14] |
| Brunello Library | Reference single-sgRNA library | Well-validated genome-wide library for performance comparisons [69] [68] |
Dual-sgRNA libraries represent a significant advancement in CRISPR screening technology, offering enhanced knockdown efficacy while reducing library size. Experimental data consistently demonstrate their superiority over single-sgRNA approaches, with 30% stronger growth suppression of essential genes and improved performance in both essentiality and drug-gene interaction screens [67] [14].
For researchers designing genetic screens, dual-sgRNA CRISPRi libraries are particularly recommended for:
- Studies requiring high knockdown efficacy without DNA damage
- Screening in complex models where library size is limiting
- Investigating essential genes and synthetic lethal interactions
- Applications where false negatives must be minimized
The field continues to evolve with emerging technologies such as qgRNA libraries showing even greater efficacy [70]. However, dual-sgRNA designs currently offer an optimal balance of performance and practicality for most screening applications. As CRISPR screening expands into more complex physiological systems, including organoids and in vivo models [71] [72], the advantages of compact, highly efficient libraries will become increasingly important for advancing functional genomics and drug discovery.
A critical challenge in functional genomics is the precise manipulation of gene expression to establish causal relationships between genes and phenotypes. Within the context of comparing overexpression versus knockdown screening research, nuclease-deficient Cas9 (dCas9) has emerged as a versatile platform for reversible and specific genetic perturbation [73] [22]. This guide provides an objective comparison of the performance of major dCas9 effector systems, supported by experimental data, to inform researchers and drug development professionals in selecting the optimal tool for their screens.
The dCas9 Platform: From Gene Editing to Programmable Regulation
The repurposing of the CRISPR-Cas9 system from a genome editor to a transcriptional regulator began with the development of catalytically dead Cas9 (dCas9). dCas9 contains mutations that inactivate its DNA cleavage ability while retaining its capacity to bind DNA in an RNA-guided manner [73]. This DNA-binding protein can then serve as a programmable scaffold, recruiting various effector domains to specific genomic loci to manipulate transcription and epigenetics without altering the underlying DNA sequence [73]. This foundational technology enables both loss-of-function (knockdown) and gain-of-function (overexpression) studies in a highly specific and multiplexable manner, addressing key limitations of earlier technologies like RNA interference (RNAi) and cDNA overexpression [22].
Comparative Performance of Major dCas9 Effector Systems
The performance of dCas9 systems varies significantly based on the effector domains fused to it. The tables below summarize the key characteristics and performance data of the primary technologies used for CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa).
Table 1: Comparison of Major dCas9 Fusion Systems for Knockdown and Activation
| System Type | System Name | Key Effector Domains | Mechanism of Action | Best Applications | Key Performance Findings |
|---|---|---|---|---|---|
| CRISPRi (Knockdown) | dCas9-KRAB | Krüppel-associated box (KRAB) domain (e.g., from KOX1) [17] | Recruits repressive complexes to promote heterochromatin formation [22]. | Genome-wide loss-of-function screens, essential gene studies [74]. | Effective repression, but performance can be variable across cell lines and sgRNAs [17]. |
| CRISPRi (Knockdown) | Novel Repressor Fusions (e.g., dCas9-ZIM3(KRAB)-MeCP2(t)) | Potent KRAB domain (ZIM3) combined with a truncated MeCP2 repressor [17]. | Combines multiple, strong repressive mechanisms for enhanced silencing. | Studies requiring high-efficiency, consistent knockdown with lower sgRNA-dependent variability. | ~20-30% better gene knockdown compared to dCas9-ZIM3(KRAB) alone; improved reproducibility across cell lines [17]. |
| CRISPRa (Activation) | dCas9-VP64 | Four tandem copies of the VP16 activation domain (VP64) [75]. | Recruits minimal transcriptional machinery to the promoter. | Experiments requiring modest gene activation. | Considered a "first-generation" activator with ~2-fold activation on average; simple but less potent [75]. |
| CRISPRa (Activation) | SAM (Synergistic Activation Mediator) | dCas9-VP64 + engineered sgRNA scaffolds recruiting MS2-p65-HSF1 [75]. | Uses an RNA scaffold to recruit multiple, synergistic activation domains. | Single-gene activation where maximum expression is critical. | Consistently shows some of the highest activation levels for single genes [75]. |
| CRISPRa (Activation) | SunTag | dCas9 fused to a peptide array, recruiting multiple copies of scFv-VP64 [75]. | Uses a protein scaffold to recruit multiple copies of an activator. | Multiplexed gene activation studies. | Better than dCas9-VP64; shows activation levels lower than SAM but comparable to VPR in multiplexing [75]. |
| CRISPRa (Activation) | VPR | Direct fusion of dCas9 to a tripartite activator (VP64 + p65 + Rta) [75]. | A single fusion protein delivering three different strong activation domains. | A balanced choice for robust activation without a complex multi-component system. | Significantly higher activation than dCas9-VP64; generally lower than SAM but with simpler delivery [75]. |
Table 2: Quantitative Performance Summary of dCas9 Effectors
| System | Target | Cell Line | Reported Efficacy / Fold-Change | Key Advantage | Key Limitation |
|---|---|---|---|---|---|
| dCas9-ZIM3-MeCP2(t) (CRISPRi) | Endogenous Gene Transcripts | Multiple human cell lines | Improved repression at transcript and protein level [17] | High efficiency, lower variability | Larger fusion protein size |
| dCas9-VP64 (CRISPRa) | Reporter & Endogenous Genes | HEK293T, K562 | ~2-fold activation [75] | Simplicity of construct | Modest activation levels |
| SAM (CRISPRa) | Endogenous Genes | K562, A375 | Very high activation (among the highest) [75] | Maximum activation potency | Requires engineered sgRNA |
| VPR (CRISPRa) | Endogenous Genes | K562, A375 | High activation (lower than SAM) [75] | Strong activation from a single fusion | Larger construct size |
| SunTag (CRISPRa) | Endogenous Genes | K562, A375 | High activation (comparable to VPR) [75] | Amplified recruitment of activators | Relies on large antibody chains |
Experimental Insights and Protocols for Validation
A critical step in implementing these tools is the validation of their efficacy. The following are key methodologies used to characterize dCas9 fusion proteins.
Characterizing DNA Binding and Repression Efficiency
A common method to test novel dCas9 repressors, as described in a 2025 study, involves a reporter assay in human cell lines [17].
- Protocol: An eGFP reporter gene under the control of a constitutively active promoter (e.g., SV40) is introduced into cells (e.g., HEK293T) along with the dCas9-effector construct and a sgRNA targeting the promoter. The level of transcriptional repression is then quantified by measuring the reduction in eGFP fluorescence using flow cytometry [17]. This method allows for high-throughput screening of dozens of repressor domain combinations.
Validating Binding Affinity with uniBAss ELISA
The uniBAss (uniCAS Binding Assay) is an ELISA-based method developed to biochemically characterize the DNA-binding capacity of dCas9 fusion proteins [76].
- Protocol:
- A 96-well plate is coated with streptavidin, followed by biotinylated double-stranded oligonucleotides containing the target DNA sequence.
- Cell lysates containing the HA-tagged dCas9 fusion protein, pre-complexed with its crRNA, are applied to the wells.
- After washing, a primary mouse anti-HA antibody is added to detect the bound dCas9.
- A secondary anti-mouse antibody conjugated to horseradish peroxidase (HRP) is used for colorimetric detection (e.g., with ABTS).
- The absorption at 405 nm is measured, which corresponds to the amount of dCas9 bound to its target [76]. This data can be normalized to total dCas9 protein levels from a western blot to compare the intrinsic binding properties of different dCas9 fusions, independent of their expression levels [76].
Diagram 1: Core mechanism of dCas9-effector function.
Application in Genetic Screens: Knockdown vs. Overexpression
The choice between CRISPRi (knockdown) and CRISPRa (overexpression) screens is dictated by the biological question. The orthogonal insights they provide are powerful for defining gene function.
CRISPRi Knockdown Screens are ideal for identifying essential genes, vulnerabilities in cancer cells, and genes whose loss confers resistance to a drug or toxin [22]. A key advantage of CRISPRi over nuclease-based knockout (CRISPRn) is its reversibility and the avoidance of DNA damage, which can confound phenotypic readouts [17] [22]. Furthermore, CRISPRi is the preferred tool for studying the function of non-coding RNAs (lncRNAs) [22] [74].
CRISPRa Overexpression Screens are powerful for discovering genes that, when upregulated, drive resistance to drugs, promote cell growth, or induce differentiation [22]. They are particularly useful for identifying tumor suppressor genes and for probing the function of genes that are normally silent in the cell type of interest, as these would not produce a phenotype in a loss-of-function screen [22].
Diagram 2: Selecting screening approaches based on research goals.
Table 3: Key Research Reagent Solutions for dCas9 Experiments
| Item | Function in Research | Example Application in Protocols |
|---|---|---|
| dCas9-Effector Plasmid | Expresses the core dCas9 protein fused to the effector domain (e.g., KRAB, VP64, VPR). | Base vector for all experiments; requires stable cell line generation or transient transfection [17] [74]. |
| Lentiviral sgRNA Library | Delivers a pooled collection of sgRNAs for high-throughput screening. | Used in pooled screens to transduce cells at low MOI, ensuring one sgRNA per cell [22] [74]. |
| Reporter Plasmid (e.g., eGFP) | Provides a quantifiable readout for initial testing and optimization of dCas9-effector systems. | Used in reporter assays (e.g., SV40-eGFP) to measure repression/activation efficiency via flow cytometry [17]. |
| HA-Tag Antibody | Enables detection and immunoprecipitation of tagged dCas9 fusion proteins. | Critical for assays like uniBAss ELISA and western blot normalization to quantify binding and expression [76]. |
| Optimized Buffer Formulations | Maintains proper ionic conditions for dCas9 binding in in-vitro assays. | uniBAss protocol uses Tris-based buffer with optimized MgCl₂ (e.g., 5 mM) and NaCl (e.g., 90 mM) concentrations [76]. |
The landscape of dCas9 effector proteins offers a diverse toolkit for precision genetic control. For knockdown screens, novel combinatorial repressors like dCas9-ZIM3(KRAB)-MeCP2(t) set a new standard for efficiency and consistency. For activation screens, the choice between highly potent but complex systems like SAM and robust, simpler systems like VPR depends on the required level of overexpression and experimental constraints. By aligning the specific strengths—including efficacy, simplicity, and reliability—of each dCas9 fusion system with the goals of the functional genomic screen, researchers can optimally leverage these powerful tools to dissect genetic networks and accelerate therapeutic discovery.
In the comparison of overexpression versus knockdown screens, a critical methodological challenge emerges: the phenomenon of functional escape, wherein cells bypass genetic perturbations to express functional gene products. This limitation profoundly impacts the validation of gene function and therapeutic target identification. Recent research reveals that conventional single-exon targeting approaches frequently produce truncated protein isoforms through alternative splicing, compromising experimental integrity and potentially leading to erroneous conclusions in functional genomics [31] [77]. This technical guide systematically compares contemporary methodologies to overcome functional escape, focusing on strategic exon targeting approaches that enhance the reliability of knockdown constructs for research and drug development applications.
The fundamental problem stems from cellular adaptation mechanisms. When researchers target a single exon using CRISPR/Cas9 or RNAi systems, cells can exploit natural splicing alternatives to skip the targeted exon, joining upstream and downstream exons to create in-frame transcripts that yield partially functional proteins [77]. Studies characterizing mutant mouse strains generated through different gene-targeting strategies found that at least 4% exhibit conflicting phenotypes due to such adaptation mechanisms [77]. This biological resilience necessitates more sophisticated genetic intervention strategies, particularly for genes with multiple isoforms or redundant functional domains.
Molecular Mechanisms of Functional Escape
Exon Skipping and Translation Reinitiation
The primary mechanism enabling functional escape involves exon skipping events that bypass disrupted genomic regions. Comprehensive analysis of Rhbdf1 mutant mouse strains demonstrated that targeted KO-first and CRISPR/Cas9 approaches frequently result in gain-of-function alleles rather than the intended null alleles, as mutant transcripts reinitiate translation from downstream start codons [77]. Similarly, systematic analysis of CRISPR/Cas9-induced frameshift mutations revealed that approximately one-third of genes can generate truncated proteins through translation reinitiation or targeted exon skipping [77].
The biological context significantly influences escape frequency. Genes with multiple isoforms and those containing redundant functional domains present particular challenges. The robustness of mRNA surveillance mechanisms, including nonsense-mediated decay (NMD), further complicates knockout efficiency, as cells can differentially regulate the degradation of mutant transcripts based on specific sequence contexts [78]. The exon junction complex (EJC), comprising core proteins RBM8A, MAGOH, and eIF4A3, plays a crucial role in this process by marking exon-exon junctions and recruiting NMD factors when premature termination codons are detected [78].
Limitations of Conventional Targeting Approaches
Traditional gene disruption strategies exhibit systematic vulnerabilities to functional escape:
- CRISPR/Cas9 exon targeting: Conventional approaches often focus on early exons containing the start codon, but this can trigger in-frame splicing between flanking exons, producing residual functional protein [31] [77].
- RNA interference (RNAi): siRNA and shRNA methodologies effectively reduce mRNA levels but cannot completely eliminate expression, allowing low-level protein production that may retain biological activity [79].
- KO-first strategies: These versatile approaches enable tissue-specific or temporal gene disruption but may preserve regulatory elements that facilitate functional isoform expression [77].
Table 1: Functional Escape Mechanisms Across Gene Targeting Technologies
| Targeting Method | Primary Escape Mechanism | Functional Consequence | Reported Frequency |
|---|---|---|---|
| Conventional CRISPR/Cas9 | Exon skipping; Translation reinitiation | Truncated functional proteins | ~30% of genes [77] |
| RNA Interference (RNAi) | Incomplete mRNA suppression | Residual protein expression | Variable; dose-dependent |
| KO-First Strategy | Alternative promoter usage; Incomplete transcription termination | Functional reporter-tagged mutations | ≥4% of mouse models [77] |
| Definitive-Null Design | Complete gene deletion | Minimal escape potential | Theoretical null |
Strategic Exon Targeting Methodologies
Dual-Exon Targeting Strategy
A sophisticated approach to minimize functional escape involves simultaneous targeting of multiple exons to disrupt both the initiation codon and essential protein domains. In colorectal cancer research focusing on EpCAM, investigators engineered a novel CRISPR/Cas9 strategy that concurrently targets exons 1 and 3 [31]. This dual-exon targeting strategy effectively disrupts the initiation codon in exon 1 while simultaneously compromising the core structural domain in exon 3, theoretically preventing generation of any functional escape variants [31].
The experimental implementation of this approach demonstrated remarkable efficacy. The EpCAM knockdown vector (pGMC-KO-EpCAM) was constructed with sgRNAs designed against both exons, incorporating NotI and EcoRI restriction sites for cloning [31]. In functional validation, the dual-exon targeting approach achieved a dramatic reduction to 4% EpCAM-positive cells in HRT-18-KD-EpCAM-3 cells compared to 15% in wild-type controls (p<0.001) [31]. This represented a significant improvement over conventional single-exon targeting, confirming the strategic advantage of multi-exon disruption.
Exon-Exon Junction Targeting for Isoform Specificity
For genes with multiple isoforms, exon-exon junction targeting offers precise control over isoform-specific knockdown. This approach exploits unique sequences created when two exons join during splicing, enabling selective targeting of specific transcript variants without affecting others [80]. The methodology is particularly valuable for studying genes with opposing functions across different isoforms or when therapeutic targeting requires isoform discrimination.
The technical implementation typically utilizes Cas13d systems designed to recognize specific exon-exon junctions, though similar principles can be applied to CRISPR/Cas9 with appropriate guide RNA design [80]. The unified computational framework associated with this approach facilitates the design of highly specific targeting strategies that account for the complex splicing patterns observed in human genes.
Exon-Trapping Vectors for Enhanced Selection
Exon-trapping gene-targeting vectors incorporate a promoterless drug-resistance marker that is only expressed when correctly integrated within a transcribed genomic region [81]. This strategy provides powerful negative selection against random integration events while simultaneously ensuring that the resistance marker comes under the control of endogenous regulatory elements.
Advanced vector construction employs highly efficient att-mediated recombination to simplify the traditionally complex process of building exon-trapping constructs [81]. These systems can be further enhanced by incorporating a conditionally cytotoxic gene (e.g., diphtheria toxin A fragment) for additional counterselection against non-homologous integrants, significantly improving the recovery of correctly targeted clones [81].
Experimental Comparison of Targeting Strategies
Quantitative Functional Assessment
The functional consequences of strategic exon targeting were rigorously quantified in colorectal cancer models, with direct comparison between overexpression and knockdown approaches [31]. The experimental data revealed profound differences in phenotypic outcomes:
- Proliferation kinetics: EpCAM overexpression accelerated proliferation, with HT-29-OE cells showing a 20.1% increase in peak density on day 5 (30.76 ± 0.15 × 10⁴ vs. WT 25.62 ± 0.25 × 10⁴; p<0.001) [31].
- Doubling time: Conversely, dual-exon EpCAM knockdown in HRT-18 cells prolonged the doubling time by 8.8% (30.8 h vs. WT 28.3 h; p<0.05) [31].
- Migration capacity: HT-115-OE cells achieved complete scratch closure (100% vs. 74.05% in WT, p<0.001), while HRT-18-KD cells showed an 80.5% reduction (p<0.001) in Transwell migration assays [31].
Table 2: Quantitative Comparison of EpCAM Modulation in Colorectal Cancer Models
| Parameter | Overexpression (OE) | Knockdown (KD) | Statistical Significance |
|---|---|---|---|
| EpCAM-Positive Cells | 89% (vs. 12% WT) | 4% (vs. 15% WT) | p<0.001 [31] |
| Peak Cell Density (Day 5) | 30.76 ± 0.15 × 10⁴ | Not reported | p<0.001 vs. WT [31] |
| Population Doubling Time | Not reported | 30.8 h (vs. 28.3 h WT) | p<0.05 [31] |
| Scratch Assay Closure | 100% (vs. 74.05% WT) | 80.5% reduction | p<0.001 [31] |
| Transwell Migration | Hierarchy: HT-29-OE > HT-115-OE > HRT-18-KD | ANOVA p=0.0024 [31] |
Methodological Workflows for Knockdown Construct Development
The successful implementation of strategic exon targeting requires meticulous experimental workflows. The following diagram illustrates the key decision points in constructing knockdown vectors resistant to functional escape:
Comparative Analysis of Validation Methodologies
Robust validation is essential to confirm successful genetic perturbation and rule out functional escape. The comparative effectiveness of validation methodologies was assessed across multiple studies:
Table 3: Validation Methods for Genetic Perturbation Studies
| Validation Method | Detection Target | Throughput | Key Applications | Technical Considerations |
|---|---|---|---|---|
| Flow Cytometry | Surface protein expression (e.g., CD326/EpCAM) | Medium | Quantitative measurement of protein reduction [31] | Requires specific antibodies; surface expression only |
| Quantitative Western Blotting | Protein levels with size resolution | Low | Confirm knockout at protein level; detect truncated isoforms [82] | Semi-quantitative; optimization required |
| In-Cell Western Assay | Protein levels in fixed cells | High | Functional siRNA/CRISPR screens; high consistency (Z' factor) [82] | Limited antibody availability; cultured cells only |
| RNA Sequencing | Transcriptome-wide expression and splicing | Medium | Detect aberrant splicing; comprehensive isoform analysis [77] | Costly; computational analysis required |
| Functional Proliferation Assays | Cellular growth kinetics | Low | Confirm phenotypic consequences of knockdown [31] | Time-consuming; indirect measurement |
Integrated Experimental Protocols
Dual-Exon Targeting Vector Construction
The development of the pGMC-KO-EpCAM vector exemplifies the implementation of strategic exon targeting [31]:
- sgRNA Design: Design sgRNAs using computational tools (e.g., CRISPR ERA website) targeting exons 1 and 3 with the U6 promoter serving as the driving promoter.
- Restriction Site Incorporation: During synthesis of the full-length gene DNA sequence, incorporate NotI and EcoRI restriction sites at both ends of the sgRNA cassette.
- Vector Ligation: Ligate the synthesized sequence into the empty pGMC00010 vector (Addgene) using the incorporated restriction sites.
- Bacterial Transformation: Transform E. coli DH5α with the recombinant vector and culture overnight on ampicillin-resistant LB plates at 37°C.
- Sequence Verification: Select monoclonal colonies for sequencing using primer F: GTTCGGAAACCTGATTGC.
- Plasmid Amplification: Expand confirmed colonies and extract plasmids using an endotoxin-free extraction kit before storage at -20°C.
Cell Transfection and Phenotypic Characterization
Comprehensive functional validation following transfection includes multiple orthogonal assays:
- Transfection Optimization: Transfert selected CRC cell lines (HT-29, HT-115, HRT-18) using optimized Lipofectamine 2000 transfection protocols [31].
- Surface Expression Analysis: Quantify EpCAM surface expression using flow cytometry with CD326-PE antibodies at 48-72 hours post-transfection [31].
- Proliferation Kinetics: Perform daily cell counting over 8 days to establish proliferation curves and calculate population doubling times [31].
- Migration Capacity: Conduct scratch wound healing assays (0/24/48 h) and Transwell migration assays (8-μm pores, 18 h incubation) to evaluate metastatic potential [31].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Strategic Exon Targeting Studies
| Reagent / Tool | Specific Example | Function in Experiment | Technical Notes |
|---|---|---|---|
| CRISPR Vector System | pGMC00010 empty vector [31] | Backbone for sgRNA expression | Enables stable integration; contains selection markers |
| Restriction Enzymes | NotI, EcoRI FastDigest enzymes [31] | Vector linearization and insert digestion | Ensure compatible ends; prevent self-ligation |
| Transfection Reagent | Lipofectamine 2000 [31] | Nucleic acid delivery into cells | Optimize for specific cell lines; minimize toxicity |
| Selection Antibiotic | Puromycin, Neomycin [31] [81] | Selection of successfully transfected cells | Determine optimal concentration by kill curve |
| Detection Antibody | CD326(EPCAM)-PE [31] | Flow cytometry detection of surface target | Validate specificity; titrate for optimal signal |
| Endotoxin-Free Plasmid Kit | E.Z.N.A. Endo-Free Plasmid Mini Kit [31] | High-quality plasmid preparation | Critical for sensitive cell lines; improves viability |
Strategic exon targeting represents a methodological advance in genetic perturbation studies, directly addressing the critical challenge of functional escape that compromises both basic research and therapeutic development. The dual-exon targeting approach demonstrated in EpCAM research provides a robust template for optimizing knockdown constructs across diverse gene targets [31]. The consistent finding that genes adapt to single-exon targeting through exon skipping or translation reinitiation underscores the necessity of these sophisticated approaches [77].
For the research community comparing overexpression and knockdown screens, these findings highlight the importance of validation rigor in experimental design. The integration of multiple validation methods—from flow cytometry to functional phenotyping—provides essential triangulation to confirm successful genetic perturbation [31] [82]. Furthermore, the strategic selection of targeting approaches based on gene structure and isoform complexity enables researchers to maximize knockdown efficiency while minimizing confounding factors introduced by functional escape.
As genetic screening technologies continue to evolve toward higher throughput and clinical application, the principles of strategic exon targeting will play an increasingly vital role in ensuring the reliability and reproducibility of functional genomics research. The methodological framework presented here provides a foundation for developing genetically defined models that faithfully represent target gene inhibition, ultimately accelerating both basic discovery and therapeutic development.
Mitigating False Positives and Off-Target Effects in Both Modalities
Functional genetic screens using overexpression (gain-of-function) and knockdown (loss-of-function) modalities are powerful tools for elucidating gene function and identifying novel drug targets. However, each approach presents distinct challenges regarding false positives and off-target effects that can confound data interpretation. Overexpression screens may induce promiscuous interactions and stoichiometric imbalances due to non-physiological protein levels [83]. Conversely, knockdown screens, particularly those utilizing RNA interference (RNAi), are notoriously prone to off-target effects where short interfering RNAs (siRNAs) silence genes with partial sequence complementarity, leading to convoluted phenotypes [84]. Understanding the sources of these artifacts and implementing rigorous mitigation strategies is paramount for deriving biologically relevant conclusions from high-throughput screening data.
Understanding False Positives and Off-Target Effects
Overexpression Screens: Mechanisms of Artifact Generation
The artificial elevation of protein expression can disrupt cellular systems through several defined mechanisms. In absolute overexpression experiments, where a strong promoter drives expression independently of native levels, proteins with low native expression are disproportionately affected, potentially leading to massive, non-physiological overexpression and cellular defects [83]. The primary mechanisms include:
- Resource Overload: Excessive demand on cellular resources like amino acids, nucleotides, and energy can overwhelm biosynthesis and degradation pathways.
- Stoichiometric Imbalance: Disruption of multi-protein complexes occurs when one subunit is overexpressed, titrating other essential components.
- Promiscuous Interactions: At high concentrations, proteins may interact with non-cognate partners, creating false signaling pathways.
- Pathway Modulation: Artificial activation or suppression of signaling pathways can occur independently of normal regulatory constraints [83].
For example, overexpression of the P2X7 receptor was shown to promote migration and invasion in non-small cell lung cancer (NSCLC) cells by activating PI3K/Akt/GSK-3β and JNK signaling pathways [27]. While biologically informative, such effects must be distinguished from artifacts arising from the overexpression context itself.
Knockdown Screens: Mechanisms of Artifact Generation
Knockdown technologies, especially RNAi, face a different set of challenges, with off-target effects representing a major confounding factor.
- RNAi Off-Target Effects: siRNAs can silence genes with only 6-7 nucleotide complementarity to the "seed" region of the siRNA, leading to unintended phenotypic consequences [84]. It is estimated that the cumulative off-target contribution can be much higher than the on-target effect, potentially responsible for up to 30% of positives identified in a screen [84].
- Incomplete Knockdown: Partial suppression of gene expression may be insufficient to elicit a phenotype, leading to false negatives, or may produce hypomorphic phenotypes distinct from full knockout.
- CRISPR-Cas9 Off-Target Effects: While more specific than RNAi, CRISPR-Cas9 can induce double-strand breaks at genomic sites with sequence similarity to the intended sgRNA target, leading to unintended mutations and phenotypic confounders [68].
The following diagram illustrates the core mechanisms that generate artifacts in both screening modalities, highlighting the distinct pathways for false positives in each approach.
Experimental Protocols for Mitigation
Protocol for an Overexpression Screen with Built-in Controls
This protocol outlines a genome-wide ORF overexpression screen designed to identify genes conferring resistance to a lethal chemical treatment, incorporating steps to mitigate false positives [85].
- Step 1: Library Preparation. Clone a collection of human ORFs (e.g., 12,200 ORFs) into a lentiviral expression vector with an inducible promoter (e.g., tetracycline-responsive element) and a linked fluorescent reporter (e.g., Venus) [85].
- Step 2: Stable Cell Line Generation. Infect recipient cells (e.g., HEK293_M2) expressing the reverse tetracycline-controlled transactivator (rtTA) at a low multiplicity of infection (MOI ~0.3) to ensure single-copy integration. Induce expression with doxycycline and sort fluorescent cells to select for populations with functional, inducible ORF expression [85].
- Step 3: Drug Selection. Treat the pooled ORF-expressing cell population with the drug of interest at a concentration that kills >90% of control cells. Culture the surviving resistant cell pool.
- Step 4: Genomic DNA Extraction and Sequencing. Extract genomic DNA from the pre- and post-selection cell populations. Amplify the integrated ORF sequences by PCR and identify enriched ORFs via next-generation sequencing.
- Step 5: Hit Validation. Individually clone candidate ORFs and retest them for their ability to confer drug resistance in a secondary, low-throughput assay. Employ an orthogonal expression system (e.g., piggyBac transposon) to confirm phenotype independence from the screening system [85].
Protocol for a Knockdown Screen Addressing Off-Targets
This protocol describes a dual CRISPR knockout and activation screen, leveraging computational design and multiple guides to minimize off-target effects [13] [84].
- Step 1: sgRNA Library Design. Use a computationally designed library (e.g., Brunello, GeCKO) with high on-target efficiency scores. Include multiple sgRNAs (typically 4-6) per gene to control for phenotype consistency and include negative control sgRNAs with no known targets [68].
- Step 2: Lentiviral Production. Package the sgRNA library into lentiviral particles using a second- or third-generation system to produce high-titer virus.
- Step 3: Cell Transduction. Transduce the target cells at a low MOI (0.3-0.6) to ensure most cells receive a single sgRNA. Maintain a representation of at least 200-500 cells per sgRNA to prevent stochastic dropout [68].
- Step 4: Phenotypic Selection. Apply the selective pressure (e.g., ATR inhibitors VE822 or AZD6738) [13]. For resistance screens, use high drug doses to kill most cells and isolate resistant clones.
- Step 5: NGS and Hit Analysis. Extract genomic DNA from selected and control populations. Amplify the sgRNA sequences and perform NGS. Use algorithms (e.g., RSA, MAGeCK) to identify significantly enriched or depleted sgRNAs. Genes targeted by multiple, independent sgRNAs are considered high-confidence hits [13].
Comparative Data and Mitigation Strategies
The table below summarizes the primary sources of artifacts and corresponding mitigation strategies for each screening modality.
Table 1: Comparative Analysis of Artifact Sources and Mitigation in Genetic Screens
| Screening Modality | Primary Source of Artifacts | Key Mitigation Strategies | Supporting Experimental Data |
|---|---|---|---|
| Overexpression | Non-physiological protein levels causing resource overload, stoichiometric imbalance, and promiscuous interactions [83]. | • Use of inducible promoters [85].• Dose-response validation [27].• Rescue experiments (knockdown after overexpression).• Orthogonal validation (e.g., piggyBac system) [85]. | P2X7 receptor study: Overexpression promoted NSCLC cell migration/invasion; knockdown yielded opposite effects, confirming phenotype specificity [27]. |
| Knockdown (RNAi) | siRNA seed-based off-target effects; can account for ~30% of screen positives [84]. | • Use of multiple siRNAs/shRNAs per gene [68].• Computational seed analysis (e.g., TargetScan) [84].• pNEMs to model off-target effects [84].• Validation with CRISPR knockout. | pNEMs: Improved pathway reconstruction from RNAi data by explicitly modeling combinatorial knockdown probabilities from off-targets [84]. |
| Knockout (CRISPR) | Off-target DNA cleavage at genomic sites with sequence similarity to the sgRNA [68]. | • Use of optimized sgRNA libraries (e.g., Brunello) [68].• Paired nickases (Cas9 D10A mutant).• Confirmation with multiple sgRNAs per gene [13].• Dual knockout/activation screens for cross-validation [13]. | Dual CRISPR Screen: Identified ATRi resistance genes. Knockout and activation results for the same gene provided complementary, validating evidence [13]. |
The following workflow integrates these mitigation strategies into a cohesive framework for conducting a robust genetic screen, from initial design to final hit confirmation.
Successful execution of genetic screens relies on a suite of well-validated reagents and computational tools. The table below lists key resources for implementing the protocols and mitigation strategies discussed.
Table 2: Essential Research Reagents and Tools for Genetic Screens
| Resource Name | Type/Function | Specific Use Case |
|---|---|---|
| hORFeome Collection | A curated library of human Open Reading Frames (ORFs) cloned into a Gateway-compatible vector [85]. | Conducting genome-wide overexpression screens; enables systematic gain-of-function studies. |
| Brunello CRISPR Knockout Library | A genome-scale CRISPR knockout sgRNA library targeting 19,114 genes with 76,441 guides, designed for high on-target efficiency and minimal off-target effects [13] [68]. | Performing high-fidelity loss-of-function screens with reduced false-positive rates. |
| Calabrese CRISPR Activation Library | A library of sgRNAs designed to recruit transcriptional activators to gene promoters for targeted gene overexpression [13]. | Conducting gain-of-function screens without full-length ORF cloning; complementary to knockout screens. |
| TargetScan | A bioinformatics tool that predicts biological targets of miRNAs and siRNAs by searching for the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of the RNAi molecule [84]. | Predicting and analyzing potential off-target effects in RNAi screening data. |
| pc-NEMs (Probabilistic Combinatorial Nested Effects Models) | An R package for network inference that explicitly incorporates siRNA off-target prediction data to improve the reconstruction of signaling pathways from RNAi screens [84]. | Deconvoluting complex RNAi screening data by modeling combinatorial knockdowns from off-target effects. |
| Lentiviral Vectors (3rd Generation) | A safe and efficient viral delivery system for stably integrating genetic payloads (ORFs, sgRNAs) into a wide range of cell types, including non-dividing cells [68] [85]. | Delivering screening libraries into mammalian cells for stable, long-term expression of the perturbation. |
The strategic comparison between overexpression and knockdown screens reveals a complementary landscape of strengths and vulnerabilities. Overexpression screens risk artifacts from supra-physiological expression but excel in identifying drug resistance mechanisms and pathway components [27] [85]. Knockdown screens, while powerful for defining essential genes and pathways, must contend with persistent off-target effects, even with advanced tools like CRISPR [13] [84]. The most robust conclusions in functional genomics are drawn not from a single screen but from a convergent approach that leverages both modalities. Using structured experimental designs, incorporating computational corrections, and employing orthogonal validation are non-negotiable for mitigating false discoveries. By systematically understanding and addressing the unique artifact profiles of each method, researchers can harness their full power to advance drug discovery and unravel complex biological networks.
In the field of functional genomics, high-throughput perturbation screens—including both knockdown and overexpression studies—have become indispensable for deciphering gene function and identifying therapeutic targets. However, the value of these screens hinges on the rigorous evaluation of their performance. Recent studies have revealed troubling anomalies in benchmarking practices, where naive predictors like the simple mean of all perturbed cells can outperform sophisticated deep learning models on standard metrics due to systemic control biases and metric artifacts [86]. This underscores a critical need for more robust benchmarking frameworks. The choice between knockdown (typically using CRISPRi or RNAi) and overexpression (using CRISPRa) approaches introduces further complexity, as each modality presents distinct mechanistic profiles and benchmarking challenges. This guide provides a comprehensive comparison of evaluation metrics and methodologies, enabling researchers to accurately assess perturbation quality and screen performance within the broader context of comparing overexpression versus knockdown screening paradigms.
Core Metrics for Evaluating Perturbation Responses
The accurate assessment of perturbation screen outcomes requires metrics that are sensitive to true biological signals while resistant to common artifacts like control population bias.
Established Metrics and Their Limitations
Traditional metrics for evaluating perturbation responses have primarily included:
- Mean Squared Error (MSE) and Mean Absolute Error (MAE): Measure the average squared or absolute difference between predicted and observed expression values across all genes [86] [6].
- Pearson Correlation of Delta (Pearson(Δ)): Assesses the correlation between predicted and observed expression changes relative to a control mean profile [86].
However, these standard metrics have significant limitations. The mean baseline predictor—which simply predicts the average of all perturbed cells regardless of the specific perturbation—often achieves surprisingly high performance on these metrics, sometimes outperuning complex models. This occurs because control-referenced deltas and unweighted error metrics can reward "mode collapse" (predicting the same output regardless of input) when the control is biased or the biological signal is sparse [86].
Advanced Metric Frameworks
To address these limitations, researchers have developed improved metric frameworks:
- Weighted MSE (WMSE) and Weighted R²(Δ): These DEG-aware metrics measure error with respect to all perturbations (not just controls) and apply higher weights to genes that are typically differentially expressed, increasing sensitivity to niche biological signals [86].
- Calibrated Performance Baselines: Effective benchmarking requires comparison against appropriate baselines:
- Negative Baseline: The mean predictor, which should achieve null performance on properly calibrated metrics.
- Positive Baseline: A technical duplicate baseline that estimates optimal performance given the intrinsic variance of the dataset [86].
- Multi-Scale Phenotypic Metrics: For image-based screens assessing complex phenotypes like 3D genome organization, metrics must capture effects across different biological scales—from chromatin domains to nuclear architecture [87].
Table 1: Comparison of Key Metrics for Perturbation Screen Evaluation
| Metric | Calculation | Strengths | Weaknesses | Optimal Use Case |
|---|---|---|---|---|
| MSE/MAE [86] [6] | Average L2/L1 distance between predicted and observed expression | Simple, interpretable, provides absolute error magnitude | Rewards mode collapse; insensitive to sparse biological signals | Initial quality control; datasets with dense effects |
| Pearson(Δ) [86] | Correlation between predicted and observed changes from control | Captures directional agreement; controls for baseline expression | Highly sensitive to control bias; vulnerable to metric artifacts | Comparing effects of the same perturbation across systems |
| WMSE [86] | MSE weighted by likelihood of being differentially expressed | Reduces mode collapse reward; sensitive to niche signals | Requires DEG reference distribution; more complex interpretation | Primary evaluation metric; screens with sparse, strong effects |
| R²_w(Δ) [86] | Weighted variance explanation of perturbation effects | DEG-aware; measures proportion of biological signal captured | Complex interpretation; requires reference distribution | Primary evaluation metric; comparison across screens |
| Multi-Scale Phenotyping [87] | Quantitative imaging features at different biological scales | Captures complex, high-content phenotypes | Requires specialized imaging and analysis | Image-based screens (e.g., chromatin organization) |
Benchmarking Knockdown versus Overexpression Screens
The methodological choice between knockdown and overexpression significantly impacts screen design, outcome interpretation, and benchmarking approaches.
Technical Implementation and Mechanistic Differences
Knockdown (typically using CRISPRi) and overexpression (using CRISPRa) approaches employ distinct molecular mechanisms:
- CRISPRi (Knockdown): Utilizes a catalytically dead Cas9 (dCas9) fused to transcriptional repressor domains (such as KRAB or Zim3) to target gene promoters and suppress transcription [88] [67]. It does not introduce DNA double-strand breaks, avoiding genomic rearrangements and DNA damage-associated toxicity [67].
- CRISPRa (Overexpression): Employs dCas9 fused to transcriptional activators (such as VP64-p65-Rta or SunTag systems) to target gene promoters and enhance transcription [88].
Knockdown approaches typically achieve partial reduction of gene expression (often ~60-95% depending on the effector and target), while overexpression can increase transcript levels by several fold [67] [6]. This fundamental difference in the magnitude and direction of perturbation effects necessitates distinct benchmarking strategies.
Performance Considerations and Benchmarking Implications
Each approach presents unique advantages and benchmarking challenges:
Knockdown Screen Considerations:
- Enables titration of gene expression and study of essential genes through partial depletion [67].
- Produces more homogeneous loss-of-function compared to CRISPR knockout, which can generate mixed populations due to in-frame indels [67].
- Benchmarking must verify on-target efficacy while accounting for incomplete knockdown, which creates mixed populations of affected and unaffected cells [88].
- Dual-sgRNA designs can significantly enhance knockdown efficacy, with one study showing a 29% stronger growth phenotype for essential genes compared to single-sgRNA approaches [67].
Overexpression Screen Considerations:
- Particularly valuable for studying transcription factors in cell fate reprogramming or identifying oncogenes [6].
- Benchmarking requires confirmation that the targeted transcript is actually increased, with success rates ranging from 73% to over 92% across different datasets [6].
- May produce non-physiological effects due to supraphysiological expression levels, complicating biological interpretation.
Table 2: Comparison of Knockdown and Overexpression Screening Approaches
| Characteristic | Knockdown (CRISPRi) | Overexpression (CRISPRa) |
|---|---|---|
| Molecular Mechanism | dCas9 fused to repressors (KRAB, Zim3) [67] | dCas9 fused to activators (VP64-p65-Rta) [88] |
| Typical Effect Size | 60-95% reduction [67] | Several-fold increase [6] |
| Genetic Reversibility | Reversible [67] | Reversible |
| Key Applications | Essential gene study, non-coding RNA interrogation [67] | Transcription factor programming, oncogene identification [6] |
| Benchmarking Focus | On-target efficacy, incomplete knockdown effects [88] | Transcript increase verification, non-physiological effects [6] |
| Typical Efficacy Rate | Varies by effector; dual-sgRNA enhances efficacy [67] | 73-92% success in increasing target transcript [6] |
| Primary Artifacts | Variable knockdown efficiency [88] | Supraphysiological effects |
Experimental Protocols for Benchmarking
Robust benchmarking requires standardized experimental designs and data analysis protocols.
High-Content Screening Workflow
The Perturb-tracing method exemplifies an advanced workflow for image-based screening that combines pooled CRISPR screening with high-content phenotypic readouts:
Perturb-tracing screening workflow for high-content phenotyping.
This protocol enables correlating specific genetic perturbations with complex phenotypic readouts by combining several advanced technologies:
- Library Design and Cell Line Generation: A pooled sgRNA library is cloned with unique molecular barcodes and transduced into Cas9-expressing cells at low multiplicity of infection to ensure single perturbations per cell [87].
- BARC-FISH (Barcode Amplification by Rolling Circle and FISH): Linear and padlock probes hybridize to each barcode "digit," followed by circularization, ligation, and rolling circle amplification to generate strong signals for sequential fluorescent imaging and barcode decoding [87].
- Multi-Scale Phenotyping: Chromatin tracing maps 3D genome organization across multiple length scales (e.g., TADs, compartments, territories), providing high-content phenotypic profiles [87].
- Quality Control: Cell cycle staging (e.g., Geminin staining for G1 phase selection) controls for cell cycle effects on phenotypes [87].
In Vivo Perturbation Screening Protocol
Massively parallel in vivo Perturb-seq adapts perturbation screening to living organisms:
- Library Generation: sgRNA sequences targeting genes of interest are designed and cloned into AAV vector plasmids, which are pooled to create the perturbation library [88].
- AAV Production and Quality Control: AAVs containing the gRNA vector pool are produced and purified, with titer determination and NGS assessment of gRNA distribution [88].
- In Vivo Administration: AAV pools are administered via in utero injection into the lateral ventricles of developing embryos [88].
- Tissue Processing and Sequencing: Tissues are harvested, perturbed cells are enriched, and scRNA-seq libraries are prepared for data collection [88].
Computational Benchmarking Framework
For in silico perturbation prediction, the PEREGGRN platform provides a standardized benchmarking framework:
- Data Splitting: A non-standard data split ensures no perturbation condition occurs in both training and test sets, with distinct perturbation conditions allocated to test data [6].
- Perturbed Gene Handling: Directly targeted genes are set to 0 (for knockout) or observed values (for knockdown/overexpression) to avoid illusory success in prediction [6].
- Multi-Metric Evaluation: Comprehensive assessment using metrics spanning absolute error (MAE, MSE), correlation measures (Spearman), directional accuracy, and cell type classification performance [6].
Research Reagent Solutions
Successful perturbation screening depends on carefully selected reagents and tools.
Table 3: Essential Research Reagents for Perturbation Screening
| Reagent Category | Specific Examples | Function and Application | Performance Notes |
|---|---|---|---|
| CRISPR Effectors | Zim3-dCas9 [67], dCas9-KRAB [88] | Transcriptional repression for knockdown screens | Zim3-dCas9 offers excellent balance of strong on-target knockdown with minimal non-specific effects [67] |
| sgRNA Library Formats | Dual-sgRNA cassettes [67], Single-sgRNA designs | Targeting genes for perturbation | Dual-sgRNA constructs significantly improve knockdown efficacy and produce stronger phenotypes [67] |
| Viral Delivery Systems | AAV-SCH9 [88], Lentivirus | delivering perturbation constructs in vivo or in vitro | AAV-SCH9 enables rapid (48-hour) expression onset in vivo with broad cell labeling [88] |
| Barcoding Systems | BARC-FISH [87], RNA barcodes | Identifying perturbation identities in pooled screens | Ten-digit ternary barcodes enable robust decoding while compatible with chromatin tracing [87] |
| Phenotypic Assays | Chromatin Tracing [87], scRNA-seq [86] | Measuring perturbation effects at various molecular levels | Multi-scale chromatin tracing assesses 3D genome organization from TADs to nuclear architecture [87] |
Metric Implementation and Analysis Workflow
Proper implementation of benchmarking metrics requires careful consideration of their relationships and appropriate usage contexts.
Metric implementation and analysis workflow.
This workflow guides researchers through a systematic process for evaluating perturbation screens:
- Initial Assessment: Calculate traditional metrics (MSE, Pearson(Δ)) but compare performance against the mean baseline to detect potential metric artifacts [86].
- Bias Detection and Advanced Metric Application: If control bias is suspected or mode collapse is detected, implement advanced metrics like WMSE and weighted R²(Δ) that are less susceptible to these artifacts [86].
- Performance Calibration: Compare model performance against both negative (mean predictor) and positive (technical duplicate) baselines to establish realistic performance expectations [86].
- Biological Validation: Move beyond statistical measures to assess biological relevance through DEG analysis, phenotypic effect sizes, and confirmation using orthogonal experimental approaches [87] [6].
Robust benchmarking is fundamental to extracting meaningful biological insights from perturbation screens. The field is moving beyond simple correlation measures and MSE toward more sophisticated, calibrated metric frameworks that account for control biases and sparse biological signals. When comparing knockdown and overexpression approaches, researchers must select appropriate evaluation strategies that align with the distinct mechanistic profiles and expected outcomes of each modality. As perturbation technologies continue to evolve—enabling more complex phenotypic readouts and in vivo applications—benchmarking methodologies must similarly advance to ensure accurate interpretation of screening results and maintain scientific rigor in functional genomics.
Beyond the Screen: Validating Hits and Comparing Functional Outcomes
In modern drug discovery, the journey from identifying an active compound in a high-throughput screen (HTS) to confirming its biological target represents one of the most critical and challenging phases. This process, termed "hit validation," separates genuine starting points for therapeutic development from false positives that can consume extensive resources without return. Within this framework, genetic approaches—specifically CRISPR-based knockout and activation screens—have emerged as powerful complementary technologies for understanding compound mechanism of action and identifying resistance genes.
This guide examines the integrated validation workflows that leverage both chemical and genetic tools, objectively comparing their performance and applications. We focus specifically on how overexpression (CRISPRa) and knockdown (CRISPRi/knockout) screens provide distinct yet complementary insights into target biology and drug mechanism, creating a more comprehensive validation strategy than either approach alone.
Core Principles of Hit Validation
Defining Hits and Validation Criteria
Following a high-throughput screen, "hits" are typically defined as compounds that show significant activity in the primary assay [89]. However, not all hits represent genuine modulators of the intended target. The validation workflow must systematically eliminate several categories of false positives:
- Pan-assay interference compounds (PAINS) that generate signals through non-specific mechanisms [89]
- Compounds interfering with detection technology rather than the biological target
- Non-specific enzyme inhibitors such as aggregators [89]
- Compounds generating reactive products through redox cycling [89]
A well-designed validation cascade employs orthogonal assays with different readout technologies, biophysical techniques to confirm target engagement, and chemical analysis to verify compound identity and purity [89].
The Role of Genetic Screens in Validation
Genetic screens provide a powerful orthogonal approach to compound validation by identifying genes that, when modulated, alter cellular response to therapeutic compounds. Dual CRISPR knockout and activation screens enable comprehensive identification of genes that regulate resistance or sensitivity to drugs [13]. This approach allows researchers to:
- Identify genetic biomarkers of drug resistance [13]
- Uncover mechanisms of drug action through genetic modifiers
- Validate putative targets by demonstrating that genetic modulation recapitulates compound effects
- Identify potential resistance mechanisms that may emerge during treatment
Comparing Genetic Screening Approaches
CRISPR Knockout Screens
CRISPR knockout screens utilize the Cas9 nuclease to create targeted double-strand breaks in DNA, resulting in gene disruption through non-homologous end joining [13]. This approach is particularly effective for identifying genes whose loss confers resistance or sensitivity to compounds.
Key applications in validation workflows:
- Identifying synthetic lethal interactions with target pathways
- Discovering genes whose loss confers drug resistance [13]
- Mapping genetic dependencies in specific cellular contexts
Experimental protocol for genome-wide knockout screens:
- Transduce cells with a genome-wide CRISPR knockout library (e.g., Brunello library with 76,441 gRNAs targeting 19,114 genes) [13]
- Maintain 250-fold library coverage throughout the experiment
- Treat cells with compound of interest versus DMSO control
- Harvest surviving cells after appropriate treatment duration
- Extract genomic DNA and amplify gRNA sequences by PCR
- Identify enriched/depleted gRNAs by Illumina sequencing [13]
- Analyze data using algorithms like redundant siRNA activity (RSA)
CRISPR Activation Screens
CRISPR activation (CRISPRa) screens utilize a catalytically dead Cas9 (dCas9) fused to transcriptional activation domains to overexpress endogenous genes [90]. This approach enables genome-wide identification of genes whose increased expression modulates compound activity.
Key applications in validation workflows:
- Identifying genes whose overexpression confers resistance
- Discovering bypass mechanisms that overcome target inhibition
- Understanding compensatory pathways in target biology
Experimental protocol for CRISPR activation screens:
- Establish cell system with inducible dCas9-activator (e.g., in iPSC-derived microglia) [90]
- Transduce with genome-wide CRISPRa library (e.g., Calabrese library)
- Induce gene expression with doxycycline
- Apply compound selection pressure
- Harvest surviving cells and process for gRNA sequencing
- Analyze enriched gRNAs to identify resistance genes
Table 1: Comparison of Knockout vs. Activation Screening Approaches
| Parameter | CRISPR Knockout | CRISPR Activation |
|---|---|---|
| Molecular Mechanism | Cas9-induced DNA breaks cause gene disruption | dCas9-transactivator increases endogenous gene expression |
| Primary Application | Identify essential genes and loss-of-function phenotypes | Identify genes that confer phenotypes when overexpressed |
| Detection Capability | Genes whose loss causes resistance [13] | Genes whose gain causes resistance |
| Biological Insights | Reveals pathway dependencies | Reveals compensatory mechanisms and bypass pathways |
| Screen Example | ATR inhibitor resistance in HeLa cells [13] | Microglia survival and phagocytosis regulators [90] |
| Key Findings | MED12-TGFβ pathway regulates replication fork stability [13] | Disease-associated genes control microglial functions [90] |
Integrated Validation Workflows
Sequential Chemical and Genetic Validation
A robust validation workflow integrates both chemical and genetic approaches:
Phase 1: Primary Compound Validation
- Confirm primary activity in dose-response
- Test for assay interference using orthogonal formats [89]
- Assess compound behavior in ratio tests (IC50 at different enzyme concentrations) [89]
- Analyze Hill coefficients for non-specific inhibition signatures [89]
Phase 2: Target Engagement Studies
- Demonstrate direct binding using biophysical methods (SPR, DSF, ITC, MST) [89]
- Confirm binding site through structural biology (X-ray crystallography, HDX-MS) [89]
- Establish cellular target engagement using CETSA [89]
Phase 3: Genetic Validation
- Perform dual CRISPR knockout/activation screens to identify genetic modifiers [13]
- Validate individual hits using focused sgRNA libraries
- Integrate genetic findings with compound sensitivity data
Phase 4: Mechanism of Action Studies
- Determine mode of inhibition through biochemical kinetics [89]
- Assess reversibility through jump-dilution experiments [89]
- Measure binding kinetics using SPR or kinetic FRET [89]
Diagram 1: Integrated hit validation workflow combining chemical and genetic approaches
Case Study: ATR Inhibitor Resistance
A comprehensive study employing dual CRISPR knockout and activation screens identified genes regulating resistance to ATR inhibitors VE822 and AZD6738 [13]. This integrated approach revealed:
- Knockout screen findings: 393 genes (VE822) and 456 genes (AZD6738) whose loss conferred resistance, with 118 overlapping hits [13]
- Activation screen findings: Distinct sets of genes whose overexpression promoted resistance
- Biological pathways: Enrichment in protein translation, DNA replication, and sister chromatid cohesion processes [13]
- Key mechanism: MED12-mediated TGFβ pathway inhibition regulates replication fork stability [13]
This dual approach provided a more comprehensive understanding of resistance mechanisms than either method alone, highlighting how combined knockdown/overexpression screens can reveal different aspects of the same biological pathway.
Experimental Protocols and Methodologies
High-Throughput Screening Quality Control
Robust HTS requires stringent quality control measures to ensure reliable hit identification [91]:
Assay Quality Metrics:
- Z'-factor: Values between 0.5-1 indicate excellent assay quality, 0-0.5 acceptable, and below 0 unacceptable for phenotypic screens [92]
- Signal-to-background ratio: Minimum 3:1 ratio for reliable detection
- Coefficient of variation (CV): Measures dispersion of controls
- DMSO tolerance: Ensure assay performance with typical compound storage solvents
Validation protocols should include:
- Plate uniformity tests across full plate formats
- Control performance tracking across multiple runs
- Pilot screens to estimate hit rates and validate assay robustness [91]
CRISPR Screen Implementation
Library Design Considerations:
- Library coverage: Maintain ≥250-fold representation to prevent stochastic gRNA loss [13]
- gRNA design: 4-6 gRNAs per gene with optimized on-target efficiency
- Control gRNAs: Include non-targeting controls and essential/neutral gene targets
Cell Line Engineering:
- For CRISPRa/i: Stable integration of dCas9-transactivator/repressor in safe-harbor loci [90]
- Optimize induction parameters for minimal basal activity and maximal inducible expression
- Validate surface marker expression or functional responses to confirm cell type identity (e.g., microglia markers IBA1, GPR34) [90]
Table 2: Essential Research Reagents for Validation Workflows
| Reagent / Solution | Function in Validation | Example Application |
|---|---|---|
| Brunello CRISPR Knockout Library | Genome-wide loss-of-function screening | Identify genes whose loss confers drug resistance [13] |
| Calabrese CRISPR Activation Library | Genome-wide gain-of-function screening | Identify genes whose overexpression causes resistance |
| AAV Vectors (e.g., AAV7m8) | In vivo gene delivery for validation | Müller glia cell cycle reactivation in retina [93] |
| Lipofectamine 2000 | Nucleic acid transfection | EpCAM modulation in colorectal cancer cells [31] |
| CellTiter-Glo Assay | Viability readout for HTS | Measure compound cytotoxicity in phenotypic screens [92] |
| Genedata Screener | HTS data analysis | Process plate-based screening data and identify hits [91] |
| Operetta CLS + Harmony | High-content imaging and analysis | Phenotypic screening with machine learning analysis [91] |
Data Analysis and Hit Selection
HTS Data Processing:
- Normalize plate data to controls to account for inter-plate variation [92]
- Calculate Z-scores to identify outliers representing potential hits [92]
- Apply statistical cut-offs (e.g., 2-3 standard deviations from mean) for hit selection [91]
Genetic Screen Analysis:
- Map sequencing reads to gRNA library
- Calculate fold-enrichment of gRNAs in treated vs. control samples
- Use statistical algorithms (RSA, MAGeCK) to identify significantly enriched/depleted genes [13]
- Perform pathway enrichment analysis on hit genes
Diagram 2: Dual CRISPR screen analysis pathway from sequencing to validation
Comparative Performance Data
Technical Performance Metrics
Table 3: Performance Comparison of Validation Methods
| Validation Method | Throughput | Information Gained | Key Limitations | Resource Requirements |
|---|---|---|---|---|
| Orthogonal Assays | Medium-High | Confirms activity in different assay format | May miss certain false positives | Moderate (assay development) |
| Biophysical Binding | Low-Medium | Direct confirmation of target engagement | May not reflect cellular context | High (instrumentation, expertise) |
| CRISPR Knockout | High | Identifies resistance/sensitivity genes | Limited to loss-of-function effects | High (library, sequencing) |
| CRISPR Activation | High | Identifies bypass mechanisms | May produce non-physiological overexpression | High (library, sequencing) |
| Dual CRISPR Approach | High | Comprehensive genetic interaction map | Increased complexity of data integration | Very High (multiple libraries) |
Case Study Performance Data
ATR Inhibitor Screens [13]:
- Screen format: Dual genome-wide CRISPR knockout and activation
- Cell models: HeLa (cancer) and MCF10A (non-transformed)
- ATR inhibitors tested: VE822 and AZD6738
- Key results: 118 overlapping genes in knockout screens across both inhibitors
- Biological insights: Multiple resistance mechanisms including replication fork progression and apoptosis prevention
Microglia Functional Screens [90]:
- Screen format: CRISPRi/a in iPSC-derived microglia
- Phenotypes assessed: Survival, phagocytosis, activation
- Key results: Identified neurodegeneration-associated genes controlling microglia function
- Translation: Revealed microglial states matching those in human brains
Integrated validation workflows that combine chemical and genetic approaches provide the most robust path from high-throughput screening hits to confirmed targets. The complementary nature of knockdown and overexpression screens is particularly powerful, as each method reveals different aspects of cellular response to perturbation.
Key advantages of integrated approaches:
- Comprehensive coverage: Knockout screens identify essential genes while activation screens reveal compensatory mechanisms
- Pathway context: Dual screens map genetic interactions and pathway relationships
- Biomarker discovery: Identify potential resistance mechanisms before clinical development
- Target confidence: Multiple orthogonal methods increase confidence in genuine targets
As CRISPR technologies continue to evolve, we anticipate increased use of more sophisticated screening approaches, including dual gene perturbation, spatial functional genomics, and single-cell readouts. These advances will further enhance our ability to confidently navigate from initial screening hits to therapeutically relevant targets, ultimately improving the success rate of drug discovery programs.
In functional genomics, overexpression (OE) and knockdown (KD) studies represent two fundamental, yet opposing, approaches for elucidating gene function. Overexpression probes the consequences of gain-of-function (GOF), while knockdown investigates loss-of-function (LOF) phenotypes. When their results align, they provide compelling evidence for a gene's role in a biological process. When they diverge, they can reveal complex regulatory networks, buffering mechanisms, and context-specific functionalities. This guide objectively compares these methodologies, underpinned by experimental data from recent studies, to inform their strategic application in basic research and drug development.
Theoretical Foundations: A Tale of Two Perturbations
The core premise of functional genomics is that perturbing gene expression levels reveals function. Overexpression and knockdown operate from opposite ends of this spectrum.
Knockdown/Knockout approaches, particularly those utilizing CRISPR-Cas9, aim to ablates gene expression. The Cas9 nuclease introduces double-strand breaks in DNA, which are repaired by error-prone non-homologous end-joining (NHEJ), often resulting in frameshift mutations and premature stop codons that disrupt the open reading frame [94] [68]. This is ideal for identifying essential genes and pathways necessary for a phenotype.
Overexpression strategies, in contrast, increase gene dosage and activity. While traditional cDNA overexpression libraries have limitations, CRISPR activation (CRISPRa) systems have revolutionized GOF screening. Using a catalytically dead Cas9 (dCas9) fused to transcriptional activation domains like VP64, p65, and Rta (a system known as VPR), or the Synergistic Activation Mediator (SAM) complex, researchers can precisely upregulate transcription at endogenous gene loci [94]. This reveals genes whose elevated expression is sufficient to induce or modify a phenotype.
The convergence of findings from both LOF and GOF experiments provides the strongest functional validation, as demonstrated in pooled CRISPR screens that can interrogate both modalities in parallel [94] [95].
Methodological Comparison: Protocols and Workflows
The experimental workflows for overexpression and knockdown screens share a common structure but differ in their core reagents and mechanisms.
Core Workflow for Pooled CRISPR Screens
The following diagram illustrates the generalized workflow for a genome-wide pooled screen, applicable to both knockout and activation studies.
Detailed Experimental Protocols
The table below summarizes the key methodological differences in constructing and executing these experiments.
| Experimental Component | Knockdown/Knockout (LOF) | Overexpression (GOF) |
|---|---|---|
| Objective | Identify genes essential for a phenotype [68] | Identify genes sufficient to induce a phenotype [94] |
| Core Mechanism | CRISPR-Cas9 induces frameshift indels via NHEJ [94] [68] | CRISPRa (e.g., dCas9-SAM, dCas9-VPR) recruits transcriptional activators [94] |
| Guide RNA (gRNA) Library | gRNAs target early exons of coding sequences to disrupt the ORF (e.g., GeCKO, Brunello libraries) [68] | gRNAs target promoter regions near transcription start sites [94] |
| Vector Delivery | Lentiviral transduction for stable genomic integration of gRNA and Cas9/dCas9-effector [68] | Lentiviral transduction for stable genomic integration of gRNA and dCas9-activator [94] |
| Phenotypic Selection | Positive selection (e.g., drug resistance, survival) or negative selection (e.g., essential gene identification) [68] | Positive selection for induced phenotypes (e.g., proliferation, differentiation, resistance) [94] |
| Hit Identification | NGS of gRNAs from selected vs. control population; essential genes show gRNA depletion [68] | NGS of gRNAs from selected vs. control population; inducing genes show gRNA enrichment [94] |
Case Studies: Experimental Data and Convergent Insights
Direct head-to-head comparisons of overexpression and knockdown within a single study provide the most powerful evidence for gene function.
Case Study 1: EpCAM in Colorectal Cancer
A 2025 study constructed EpCAM overexpression and CRISPR/Cas9 knockdown vectors to systematically analyze its role in colorectal cancer (CRC) [31].
Experimental Protocols:
- Overexpression: The full-length human EpCAM sequence was cloned into a pCDH-CMV-MCS-EF1-RFP-T2A-puro vector using XbaI and NheI restriction sites. Plasmids were validated by restriction digestion and Sanger sequencing [31].
- Knockdown: A dual-exon targeting strategy (exons 1 and 3) was used to prevent functional escape variants. sgRNAs were cloned into a pGMC00010 vector via NotI and EcoRI sites [31].
- Cell Models: CRC cell lines (HT-29, HT-115, HRT-18) were transfected using Lipofectamine 2000. Functional assays included flow cytometry for EpCAM surface expression, cell counting for proliferation, and scratch wound healing/Transwell assays for migration [31].
Quantitative Results: The table below summarizes the key functional outcomes.
| Experimental Condition | Proliferation Impact | Migration Impact | Molecular Validation |
|---|---|---|---|
| EpCAM Overexpression (HT-29-OE) | 20.1% increase in peak cell density [31] | Enhanced scratch closure (100% vs. 74.05% in WT) [31] | 89% EpCAM-positive vs. 12% in WT [31] |
| EpCAM Knockdown (HRT-18-KD) | Doubling time prolonged by 8.8% [31] | 80.5% reduction in migration capacity [31] | 4% EpCAM-positive vs. 15% in WT [31] |
Conclusion: The convergent, opposite phenotypes from OE and KD firmly establish EpCAM as a master regulator of CRC aggressiveness, promoting both proliferation and migration [31].
Case Study 2: P2X7 Receptor in Non-Small Cell Lung Cancer
A 2025 study investigated the P2X7 receptor in NSCLC using both overexpression and knockdown approaches [27].
Experimental Protocols:
- Recombinant plasmids for P2X7 receptor overexpression and knockdown were constructed and transfected into LLC and LA795 cell lines.
- In vitro functional assays assessed migration, invasion, proliferation, and apoptosis.
- Stable cell lines were generated for in vivo tumorigenicity studies.
- Mechanistic analysis involved Western blotting for PI3K/Akt/GSK-3β, JNK signaling pathways, and epithelial-mesenchymal transition (EMT) markers [27].
Summary of Findings:
- Overexpression of P2X7 promoted migration, invasion, and tumor growth, activating oncogenic PI3K/Akt/GSK-3β and JNK pathways [27].
- Knockdown of P2X7 suppressed proliferation, promoted apoptosis, and inhibited tumor growth, suppressing the same pathways [27].
Conclusion: The convergent results from opposing manipulations identify P2X7 as a bona fide oncoprotein and a promising therapeutic target in NSCLC [27].
When Perturbations Diverge: Synergy and Complex Phenotypes
Sometimes, overexpression and knockdown are not simple opposites but are used synergistically to unlock complex phenotypes, as seen in reactivating mammalian Müller glia (MG) proliferation.
Experimental System: To overcome the quiescence of MG in the adult mouse retina, researchers simultaneously overexpressed cyclin D1 and knocked down the cell cycle inhibitor p27Kip1 using a single AAV vector [96].
Protocol:
- A single AAV7m8 vector with a GFAP promoter was used for MG-specific delivery.
- The vector co-expressed cyclin D1 and a short hairpin RNA (shRNA) targeting p27Kip1.
- Proliferation was assessed via EdU incorporation in uninjured retinas [96].
Results:
- Individual perturbations were weak: p27Kip1 knockdown alone induced minimal proliferation; cyclin D1 overexpression alone had a moderate effect.
- The combined OE/KD treatment acted synergistically, driving robust MG cell cycle re-entry without causing neoplasia. This demonstrates that some biological barriers require concurrent GOF and LOF perturbations to be overcome [96].
The synergistic relationship between these two opposing manipulations can be visualized as follows:
The Scientist's Toolkit: Essential Research Reagents
Success in functional genomics relies on a standardized toolkit of reagents and resources. The following table catalogues essential solutions for conducting head-to-head OE/KD studies.
| Research Reagent | Function & Application | Examples & Notes |
|---|---|---|
| CRISPR Libraries | Pre-designed pools of sgRNAs for genome-wide or targeted screens. | GeCKO (human/mouse) [68], Brunello (high-specificity human) [68], TKO (Toronto KnockOut) [68]. |
| Lentiviral Vectors | Efficient delivery system for stable integration of constructs into dividing and non-dividing cells. | Third-generation systems for safety; can be one-vector (Cas9+sgRNA) or two-vector systems [68]. |
| CRISPR Activators | Engineered systems for transcriptional overexpression. | SAM [94], SunTag [94], VPR [94]. Fuse dCas9 to strong activation domains. |
| Validation Assays | Confirm genetic perturbation and quantify functional effects. | Sanger Sequencing (plasmid validation) [31], Flow Cytometry (surface marker expression) [31], Scratch Wound/Transwell (migration) [31]. |
| Bioinformatic Algorithms | Identify significantly enriched or depleted gRNAs from NGS data. | MAGeCK, CERES, drugZ [68]. Correct for screen-specific biases like multiple-hypothesis testing. |
Overexpression and knockdown are complementary, not redundant, tools in the functional genomics arsenal. As the cited studies demonstrate, their convergent results provide strong validation of a gene's core function, as seen with EpCAM and P2X7. Conversely, their strategic combination can be necessary to dissect complex biological barriers and achieve therapeutic phenotypes, exemplified by the reactivation of Müller glia. The choice between—or combination of—these approaches must be guided by the biological question, with a clear understanding of the distinct but overlapping insights each method provides. The continued development of refined CRISPR tools will further enhance the precision and scope of these foundational techniques.
In functional genomics, two primary strategies—overexpression (gain-of-function) and knockdown (loss-of-function) screens—provide complementary lenses for investigating gene function and identifying therapeutic targets. Overexpression screens identify genes that, when amplified, are sufficient to induce phenotypic changes, potentially revealing novel oncogenes or resistance mechanisms [85]. Conversely, knockdown approaches determine which genes are necessary for specific biological processes, often uncovering essential genes and vulnerability factors [25]. While each method offers distinct insights, their strategic integration enables researchers to overcome the limitations inherent in either approach alone, providing a more comprehensive understanding of complex biological systems and generating more robust, physiologically relevant phenotypes.
The synergy between these modalities is particularly valuable in drug discovery and validation. As evidenced by research on ATR inhibitors, combining knockout and activation screens comprehensively identifies genetic determinants of drug resistance, revealing mechanisms that single-direction screens would miss [47] [13]. Similarly, in cancer research, simultaneous manipulation of multiple regulatory nodes—such as cyclin D1 overexpression combined with p27kip1 knockdown—can produce dramatic phenotypic changes that individual perturbations cannot achieve, demonstrating the power of coordinated genetic interventions [97]. This guide systematically compares these approaches, providing researchers with experimental frameworks and analytical tools for implementing synergistic screening strategies.
Technical Comparison: Overexpression vs. Knockdown Screens
Fundamental Principles and Mechanisms
Overexpression screens operate on the principle of gene sufficiency, introducing additional genetic material to determine whether a gene can drive a specific phenotype. Experimentally, this is achieved by transferring open reading frames (ORFs) into lentiviral expression vectors downstream of constitutive promoters, enabling ectopic gene expression in target cells [98]. The resulting phenotype indicates whether the overexpressed gene is sufficient to induce the observed effect, such as proliferation in growth-factor-deprived conditions or drug resistance.
Knockdown screens instead test gene necessity by reducing endogenous gene expression through various interference mechanisms. RNA interference (RNAi) degrades mRNA transcripts via the RNA-induced silencing complex (RISC), while CRISPR-based knockout employs Cas nucleases to create permanent, inheritable gene disruptions [25]. Morpholino oligos provide transient knockdown by blocking translation or pre-mRNA splicing without degrading targets. Each method determines whether a gene is essential for maintaining a particular cellular state or function.
Table 1: Fundamental Characteristics of Screening Approaches
| Feature | Overexpression Screening | Knockdown Screening |
|---|---|---|
| Primary Question | Is gene X sufficient to cause phenotype Y? | Is gene X necessary for phenotype Y? |
| Genetic Principle | Gain-of-function | Loss-of-function |
| Typical Approach | cDNA/ORF library expression | RNAi, CRISPR knockout, morpholinos |
| Expression Change | Increased gene product | Decreased gene product |
| Key Applications | Identifying oncogenes, drug resistance mechanisms, developmental drivers | Identifying essential genes, synthetic lethal interactions, drug targets |
| Phenotypic Outcome | Often reveals dominant effects | Can uncover recessive traits |
Experimental Platforms and Workflows
Overexpression screening platforms typically utilize lentiviral vectors for efficient gene delivery across diverse cell types. The human ORFeome collection (version 3.1 contains 12,212 ORFs representing 10,214 distinct genes, which can be transferred into destination vectors via Gateway recombination cloning [85] [98]. These systems often incorporate fluorescent reporters (e.g., IRES-controlled GFP) to monitor transduction efficiency and transgene expression, enabling high-throughput phenotypic assessment through high-content imaging or flow cytometry.
Knockdown screening methodologies have evolved from RNAi to CRISPR-Cas9 systems, offering improved specificity and efficacy. Genome-wide CRISPR knockout libraries such as Brunello target 19,114 genes with 76,441 unique guide RNAs, providing approximately four guides per gene for comprehensive coverage [47]. CRISPR activation (CRISPRa) screens represent an advanced gain-of-function approach that directly stimulates endogenous gene expression without introducing exogenous cDNA, complementing traditional knockout screens [13].
Screening Workflow Comparison: This diagram illustrates the parallel experimental pathways for overexpression and knockdown screens, culminating in integrated data analysis for synergistic insights.
Case Studies in Synergistic Screening
Dual CRISPR Screens for ATR Inhibitor Resistance
A landmark demonstration of integrated screening comes from research on ataxia telangiectasia and Rad3-related (ATR) protein kinase inhibitors. Cancer cells heavily rely on ATR to manage replication stress, making ATR inhibition a promising therapeutic strategy [47]. To comprehensively identify resistance mechanisms, researchers employed dual genome-wide CRISPR knockout and CRISPR activation screens against two distinct ATR inhibitors (VE822 and AZD6738) in both HeLa cancer cells and non-transformed MCF10A breast epithelial cells [13].
The knockout screen identified genes whose loss confers resistance, with significant overlap between both inhibitors: 118 genes were common hits between VE822 and AZD6738 screens, far exceeding random probability [47]. Seven genes (KNTC1, EEF1B2, LUC7L3, SOD2, MED12, RETSAT, and LIAS) ranked within the top 40 hits for both ATR inhibitors. Validation experiments confirmed that knockdown of each these genes significantly increased cellular resistance to ATR inhibition [47] [13]. Simultaneously, the activation screen pinpointed genes whose overexpression drives resistance, providing a complete spectrum of genetic modifiers.
Table 2: ATR Inhibitor Resistance Screening Results
| Gene | CRISPR Knockdown Phenotype | Biological Process | Validation Method |
|---|---|---|---|
| MED12 | Increased ATRi resistance | TGFβ signaling, replication fork stability | siRNA, clonogenic assays |
| KNTC1 | Increased ATRi resistance | Cell cycle regulation | Proliferation assays |
| EEF1B2 | Increased ATRi resistance | Protein translation | Multiple ATRi doses |
| SOD2 | Increased ATRi resistance | Oxidative stress response | CellTiterGlo viability |
| LUC7L3 | Increased ATRi resistance | RNA processing | Colony formation |
| RETSAT | Increased ATRi resistance | Retinol metabolism | siRNA confirmation |
| LIAS | Increased ATRi resistance | Mitochondrial function | Multiple validation assays |
Coordinated Gene Regulation in Cellular Reprogramming
Research on Müller glia cell cycle reactivation demonstrates how simultaneous knockdown and overexpression can achieve phenotypes unattainable through single interventions. In the mouse retina, neither p27Kip1 knockdown alone nor cyclin D1 overexpression alone robustly reactivated the Müller glia cell cycle [97]. However, combining both perturbations through a single AAV vector (CCA) stimulated significant proliferation and reprogramming without disrupting retinal structure or function.
This synergistic effect emerged from targeting complementary regulatory nodes: cyclin D1 overexpression promotes G1/S phase progression while p27Kip1 knockdown removes a critical cyclin-dependent kinase inhibitor [97]. The resulting phenotype exceeded additive expectations, demonstrating true synergy where the combined effect substantially surpassed the sum of individual effects. This approach successfully downregulated interferon pathway signaling, potentially contributing to retinal regeneration—a finding with significant therapeutic implications for retinal diseases.
P2X7 Receptor Modulation in NSCLC Progression
Research on non-small cell lung cancer (NSCLC) illustrates how bidirectional genetic manipulation validates therapeutic targets. Functional P2X7 receptors were expressed in LLC and LA795 NSCLC cell lines, and modulating this receptor produced diametrically opposed phenotypes depending on directionality [27]. P2X7 receptor overexpression promoted cancer cell migration, invasion, and tumor growth through PI3K/Akt/GSK-3β and JNK signaling pathways, while receptor knockdown suppressed proliferation, promoted apoptosis, and inhibited tumor growth [27].
This bidirectional validation confirmed P2X7 receptor's potential as a therapeutic target for NSCLC. The consistent inverse phenotypes across in vitro and in vivo models demonstrated the robustness of the findings, while mechanistic studies identified specific signaling pathways (PI3K/Akt/GSK-3β, JNK, and epithelial-mesenchymal transition) mediating these effects [27].
Bidirectional Target Validation: P2X7 receptor modulation in NSCLC demonstrates how overexpression and knockdown produce inverse phenotypes, strengthening therapeutic target validation.
Experimental Design and Protocols
Dual CRISPR Screening Methodology
Library Design and Coverage: For genome-wide knockout screens, the Brunello CRISPR knockout library provides comprehensive coverage with 76,441 unique gRNAs targeting 19,114 genes [47]. To maintain library representation, infect cells at a scale ensuring 250-fold coverage (approximately 20 million library-infected cells). For CRISPR activation screens, the Calabrese CRISPR activation library enables systematic gene overexpression [13]. Conduct screens in biological replicates with appropriate controls (e.g., non-targeting gRNAs).
Dosing Strategy: Unlike sensitivity screens that use low drug concentrations, resistance screens require high doses that kill most cells (e.g., 90% cell death over 108 hours) to enrich for resistant populations [47]. For ATR inhibitors, use 1.5μM VE822 or 3.6μM AZD6738 versus DMSO control. Treat cells for sufficient duration to allow gRNA enrichment or depletion—typically 10-14 days or approximately 5-6 cell doublings.
Hit Identification: Extract genomic DNA from surviving cells and amplify gRNA regions for Illumina sequencing. Analyze sequencing data using the redundant siRNA activity (RSA) algorithm to identify significantly enriched or depleted gRNAs [47]. Compare gRNA abundances between treatment and control conditions, with genes ranked by statistical significance (logP values). Validate top hits using individual gRNAs or siRNAs in secondary assays.
Lentiviral Overexpression Screening Protocol
ORF Library Management: Utilize the human ORFeome collection (version 3.1 contains 12,212 ORFs representing 10,214 genes) [85]. Divide the collection into manageable minipools (e.g., 34 pools of 376 ORFs each). Perform Gateway LR recombination to transfer ORFs en masse from pDONR223 entry vectors into lentiviral expression vectors containing constitutive promoters and fluorescent reporters.
Viral Production and Transduction: Produce lentivirus by co-transfecting expression plasmids with packaging plasmids (psPAX2 and pMD2.G) into HEK293T cells using transfection reagents like FuGENE [85]. Transduce target cells at low multiplicity of infection (MOI ≈ 0.3) to ensure single-copy integration. Use fluorescence-activated cell sorting to select successfully transduced cells based on linked reporter expression (e.g., GFP).
Phenotypic Assessment: For proliferation screens, employ nucleotide analogue incorporation (e.g., EdU) to detect DNA synthesis in transduced cells [98]. Use high-content imaging systems for automated quantification. In drug resistance screens, apply lethal compound concentrations and identify ORFs enriched in surviving populations through PCR amplification and next-generation sequencing of integrated ORFs.
Research Reagent Solutions
Table 3: Essential Research Tools for Combined Screening Approaches
| Reagent/Tool | Function | Application Examples |
|---|---|---|
| Brunello CRISPR Knockout Library | Genome-wide gene knockout | Identifying genes whose loss confers drug resistance [47] |
| Calabrese CRISPR Activation Library | Genome-wide gene activation | Identifying genes whose gain confers drug resistance [13] |
| Human ORFeome Collection (v3.1) | 12,212 human ORFs for overexpression | Functional screening of gene gain-of-function [85] [98] |
| Gateway Cloning System | High-efficiency ORF transfer between vectors | Lentiviral library construction [98] |
| Lentiviral Expression Vectors | Stable gene delivery to diverse cell types | Both overexpression and knockdown screens [85] [98] |
| psPAX2/pMD2.G Packaging Plasmids | Lentiviral particle production | Generating screening libraries [85] |
| Redundant siRNA Activity (RSA) Algorithm | Hit identification from screening data | Statistical analysis of gRNA enrichment [47] |
Analytical Framework for Synergistic Phenotypes
Defining and Quantifying Synergy
Genetic synergy occurs when combined perturbations produce phenotypic effects exceeding expectations from individual effects. Statistical frameworks for quantifying synergy include Loewe additivity (expected response if agents share molecular targets), Bliss independence (multiplicative probability for independent targets), and Highest Single Agent (HSA) approaches [99]. The HSA method defines synergy as combination effects exceeding the better of individual perturbations, providing a conservative synergy estimate.
In therapeutic contexts, synergistic selectivity measures whether drug combinations show improved specificity for disease-relevant versus control phenotypes. The selectivity index (SI) quantifies potency differences between test and control systems, while differential selectivity (ΔSI) compares combination versus single-agent selectivity [99]. Significant positive ΔSI values indicate therapeutic synergy with reduced off-target effects.
Integrated Data Analysis Strategies
Combining overexpression and knockdown data requires analytical approaches that handle multi-modal genetic evidence. Pathway enrichment analysis identifies biological processes consistently affected by both modalities, increasing confidence in their functional importance. For ATR inhibitor resistance, both screening approaches implicated protein translation, DNA replication, and sister chromatid cohesion processes [47].
Concordance scoring systems prioritize genes showing inverse phenotypes in overexpression versus knockdown conditions—for example, genes whose overexpression confers resistance while knockdown increases sensitivity. This bidirectional validation strongly supports therapeutic relevance, as demonstrated with P2X7 receptor in NSCLC [27]. Multi-phenotype integration methods, such as maximally heritable (MaxH) phenotype construction, combine correlated phenotypes to enhance statistical power in genetic association studies [100].
Combining overexpression and knockdown modalities provides a powerful strategy for uncovering robust, physiologically relevant phenotypes that single-direction screens often miss. The synergistic integration of these approaches reveals complementary genetic information, strengthens target validation, and identifies context-specific biological mechanisms. As screening technologies advance—particularly with the refinement of dual CRISPR platforms—researchers can increasingly implement these integrated frameworks to accelerate therapeutic discovery and optimize personalized medicine strategies. The experimental protocols and analytical frameworks presented here provide a foundation for designing synergistic screening campaigns that maximize biological insights while minimizing methodological artifacts.
In the field of functional genomics, two powerful approaches—overexpression and knockdown screens—enable researchers to systematically identify genes that control fundamental biological processes like cell cycle regulation. Overexpression screens identify genes that, when amplified, drive phenotypic changes, potentially revealing oncogenes and positive cell cycle regulators [50]. Conversely, knockdown or knockout screens identify genes that suppress phenotypes when suppressed, often uncovering tumor suppressors and cell cycle inhibitors. This case study examines a compelling intersection of these approaches, where researchers simultaneously overexpressed cyclin D1 (a positive cell cycle regulator) and knocked down p27Kip1 (a cell cycle inhibitor) to reactivate the cell cycle in mammalian Müller glia (MG) cells [93] [101]. This combined strategy demonstrates remarkable synergy, providing a template for how combinatorial genetic interventions can achieve what single approaches cannot, with significant implications for regenerative medicine and cancer research.
Experimental Comparison: Single vs. Combinatorial Interventions
Quantitative Comparison of Proliferation Outcomes
The efficacy of cyclin D1 overexpression and p27Kip1 knockdown was rigorously tested both individually and in combination. The quantitative data from these experiments, which clearly demonstrates the synergistic effect of the dual approach, is summarized in the table below.
Table 1: Proliferation Outcomes of Single and Combinatorial Interventions in Mouse Müller Glia
| Experimental Condition | Proliferation Outcome (EdU+ MG Cells) | Key Findings |
|---|---|---|
| Control (Non-target shRNA) | No proliferation detected [93] | Served as a baseline, confirming the quiescent state of MG in uninjured retina. |
| p27Kip1 Knockdown (KD) alone | Small number of proliferating cells [93] | Demonstrated that suppressing the inhibitor alone is insufficient for robust reactivation. |
| Cyclin D1 Overexpression (OE) alone | 3-fold increase in proliferation vs. p27Kip1 KD [93] | Showed that forcing cell cycle entry is more effective than removing an inhibitor. |
| Cyclin D1 OE + p27Kip1 KD | Robust, synergistic proliferation [93] | Combined approach achieved significant MG cell cycle re-entry, surpassing the effect of either single intervention. |
Detailed Experimental Protocol
The experimental workflow and key reagents used in this study provide a reproducible template for similar combinatorial screens.
Table 2: Key Research Reagents and Experimental Tools
| Reagent/Tool | Function/Description | Experimental Role |
|---|---|---|
| AAV7m8 Serotype | Adeno-associated virus vector for gene delivery [93]. | Ensured efficient transduction of retinal Müller glia cells. |
| GFAP Promoter | Human glial fibrillary acidic protein gene promoter [93]. | Drove Müller glia-specific expression of transgenes. |
| Cyclin D1 (Ccnd1) | G1-phase cyclin, key positive regulator of cell cycle progression [93]. | Overexpressed to push quiescent MG into the cell cycle. |
| p27Kip1 shRNA | Short hairpin RNA for gene knockdown [93]. | Targeted degradation of p27Kip1 mRNA to remove cell cycle inhibition. |
| EdU (5-ethynyl-2’-deoxyuridine) | Thymidine analog [93]. | Incorporated into DNA during S-phase to label proliferating cells. |
Methodology Summary:
- Vector Construction: A single AAV vector was engineered containing both the cyclin D1 transgene for overexpression and the p27Kip1-specific shRNA for knockdown, both under the control of the MG-specific GFAP promoter [93].
- In Vivo Delivery: The AAV vector was delivered via intravitreal injection into the eyes of postnatal day 6 (P6) C57BL/6 mice [93].
- Proliferation Labeling: To label cells that re-entered the cell cycle, mice received intraperitoneal injections of EdU for five consecutive days (from P13 to P17) [93].
- Tissue Analysis: Retinas were harvested and analyzed using immunohistochemistry to detect EdU incorporation and cell-specific markers (e.g., Sox9 for MG). The number of EdU+/Sox9+ cells was quantified to measure proliferation efficacy [93].
- Advanced Profiling: Single-cell RNA sequencing (scRNA-seq) was performed on treated retinas to analyze transcriptomic changes and fate of the proliferating MG and their daughter cells [93] [101].
Figure 1: Experimental workflow for combinatorial cell cycle reactivation, showing the key steps from viral vector construction to final analysis.
Underlying Molecular Mechanism and Signaling Pathways
The molecular rationale for this combinatorial approach lies in the complementary roles of cyclin D1 and p27Kip1 at the G1 restriction point. In quiescent mammalian Müller glia, the cell cycle is arrested due to low levels of cyclin D1 and high levels of the CDK inhibitor p27Kip1 [93]. Cyclin D1, when overexpressed, binds to and activates CDK4/6, leading to hyperphosphorylation of the retinoblastoma (pRb) protein. Phosphorylated pRb releases E2F transcription factors, which then activate genes required for S-phase entry. However, p27Kip1 acts as a potent brake on this process by binding to and inhibiting cyclin D-CDK4/6 complexes. Knocking down p27Kip1 removes this inhibition, allowing the overexpressed cyclin D1-CDK complexes to function with maximal efficiency, creating a strong, synergistic push for the cell to bypass the G1/S checkpoint [93] [102]. This mechanism is illustrated in the pathway diagram below.
Figure 2: Molecular mechanism of synergistic cell cycle reactivation. Cyclin D1 overexpression and p27Kip1 knockdown work together to forcefully drive G1/S transition.
Functional and Phenotypic Outcomes
The synergistic intervention yielded several critical outcomes beyond simple proliferation metrics:
- Robust but Self-Limiting Proliferation: The induced proliferation was potent, with a significant number of MG re-entering the cell cycle. Crucially, this proliferation was self-limiting; the MG underwent typically one round of division rather than exhibiting uncontrolled growth or leading to retinal neoplasia [93] [101].
- Transcriptomic Reprogramming: scRNA-seq analysis revealed that cell cycle reactivation led to significant changes in the MG transcriptome, including suppression of interferon signaling, activation of reactive gliosis genes, and downregulation of mature glial genes, indicating a partial dedifferentiation [93] [101].
- Latent Neurogenic Potential: While the majority of MG daughter cells retained their glial fate, expanding the MG pool, a small subset (approximately 1%) began expressing markers for retinal interneurons, such as the bipolar cell marker Otx2 and the amacrine cell marker HuC/D. This suggests the intervention unlocked a latent neurogenic potential in a small population of MG [93].
- Preservation of Retinal Integrity: Importantly, this robust cell cycle reactivation did not disrupt the overall retinal structure or deplete the essential MG population, and it did not impair retinal function, highlighting the safety and potential therapeutic value of the approach [93].
Discussion: Implications for Research and Therapy
Comparison with Other Cell Cycle-Targeting Strategies
The success of the cyclin D1/p27Kip1 strategy can be better appreciated when contrasted with other common approaches in cell cycle manipulation, as detailed in the table below.
Table 3: Comparison of Cell Cycle Reactivation Strategies
| Research/Intervention Focus | Mechanism of Action | Key Outcomes & Context |
|---|---|---|
| Cyclin D1 OE + p27Kip1 KD (This Study) | Directly manipulates downstream cell cycle engine and brake [93]. | Synergistic, robust, and safe proliferation in Müller glia; latent neurogenesis. |
| Upstream Pathway Activation (e.g., Wnt/β-catenin) | Activates signaling cascades that indirectly influence cyclin expression [93]. | Can induce proliferation but may have pleiotropic effects; less targeted. |
| Transcription Factor Reprogramming (Ascl1, etc.) | Directly reprograms cell identity towards progenitors/neurons [93]. | Can induce neurogenesis but risks depleting the essential glial population. |
| CDCA3 Overexpression (Oral Cancer) | Promotes G1/S progression by degrading CDK inhibitors [102]. | Oncogenic: drives uncontrolled proliferation and oral cancer progression. |
| ADAM28 Overexpression (Prostate Cancer) | Metalloproteinase that promotes proliferation and migration [24]. | Oncogenic: enhances cancer cell aggressiveness; a potential therapeutic target. |
Broader Implications for Overexpression/Knockdown Screens
This case study elegantly demonstrates a core principle in functional genomics: that combinatorial screens can reveal powerful synergisms that single-gene approaches miss. While overexpression of cyclin D1 alone was more effective than p27Kip1 knockdown alone, neither achieved the robust, controlled reactivation seen with the dual approach. This underscores that cellular states like quiescence are maintained by multiple, redundant mechanisms, and overcoming them often requires multi-pronged interventions.
Furthermore, the study highlights the critical importance of context. The same molecular maneuver—forcing cell cycle entry—can be therapeutic (as in regenerative contexts for post-mitotic tissues) or pathogenic (as seen with CDCA3 or ADAM28 in cancers) [24] [102] [103]. This duality makes the players identified in such screens attractive targets for both regenerative medicine and oncology. The concept of "synthetic dosage lethality," where overexpression of one gene creates a vulnerability to the inhibition of another, as explored in yeast and cancer models [50], further illustrates the therapeutic potential of leveraging genetic overexpression data.
This case study on the synergistic reactivation of the cell cycle in Müller glia provides a compelling template for the power of combinatorial overexpression and knockdown strategies. By simultaneously targeting a key positive regulator (cyclin D1) and a major inhibitor (p27Kip1), researchers achieved a robust, self-limiting proliferation that single interventions failed to produce. The detailed experimental protocol, quantitative data, and mechanistic insights offer a valuable resource for researchers designing similar screens in other systems. The findings reinforce that the future of manipulating complex cellular phenotypes, whether for regenerative purposes or to combat diseases like cancer, lies in strategically combining targeted genetic interventions based on a deep understanding of the underlying regulatory networks.
In functional genomics, resistance and sensitivity phenotypes represent two fundamental outcomes in genetic screening experiments. Resistance occurs when a genetic perturbation—such as gene knockout or overexpression—enables cells to survive better under selective pressure, while sensitivity appears when perturbations make cells more vulnerable to these pressures. Understanding the distinction between these opposing phenotypes is crucial for interpreting genetic screens accurately, especially when comparing knockdown and overexpression approaches. This guide examines what these phenotypes reveal about gene function and how to design experiments to detect them effectively, providing researchers with a framework for selecting appropriate screening strategies based on their specific biological questions.
Defining the Phenotypes: Resistance and Sensitivity
Core Concepts and Biological Significance
In genetic screens, resistance refers to a phenotype where a genetic perturbation results in improved cellular fitness or survival under specific selective conditions. Conversely, sensitivity describes a phenotype where a perturbation leads to reduced fitness or increased vulnerability to selective pressure [104]. These opposing phenotypes provide complementary insights into gene function and biological pathways.
The selective pressure applied during screening dramatically influences which phenotype emerges. Resistance screens typically apply high drug pressure (70-90% growth inhibition) to create conditions where only cells with protective genetic perturbations survive and proliferate [104]. Sensitivity screens use lower drug pressure (10-30% growth inhibition) to identify genetic perturbations that enhance a drug's effect, causing specific cell populations to drop out [104]. This fundamental difference in selection pressure means that the same genetic perturbation might yield different—even opposite—phenotypes depending on screen design.
Experimental Evidence of Opposing Phenotypes
The P2X7 receptor study provides a clear example of how opposing phenotypes emerge from different manipulation approaches. Overexpression of the P2X7 receptor promoted migration and invasion in non-small cell lung cancer (NSCLC) cells, demonstrating how increased gene activity can drive pathological processes [27]. Conversely, knockdown of the same receptor yielded the opposite effect—suppressing tumor growth, invasion, and migration [27]. This opposing relationship confirmed P2X7's role as a potential therapeutic target in NSCLC.
Mechanistically, these opposing phenotypes involved regulation of key signaling pathways. Both overexpression and knockdown experiments demonstrated involvement of the PI3K/Akt/GSK-3β pathway, JNK signaling, and epithelial-mesenchymal transition (EMT) [27], revealing the comprehensive molecular network through which P2X7 receptor expression influences cancer progression.
Table 1: Key Characteristics of Resistance and Sensitivity Phenotypes
| Characteristic | Resistance Phenotype | Sensitivity Phenotype |
|---|---|---|
| Cellular Fitness | Improved survival/proliferation | Reduced survival/proliferation |
| Selection Pressure | High (70-90% GI) | Low (10-30% GI) |
| Enrichment Pattern | Positive selection (enrichment) | Negative selection (depletion) |
| Primary Screening Goal | Identify genes conferring protection | Identify genes enhancing vulnerability |
| Typical Therapeutic Application | Understanding resistance mechanisms | Identifying synergistic targets |
Experimental Approaches: Knockdown vs. Overexpression Screens
Screening Strategies and Methodologies
Knockdown and overexpression screens represent complementary approaches for probing gene function. Knockdown screens (using CRISPRko or CRISPRi) identify what happens when gene function is reduced or eliminated, while overexpression screens (using CRISPRa) reveal consequences of increased gene activity [104] [13]. The dual genome-wide CRISPR knockout and activation screen for ATR inhibitor resistance exemplifies how these approaches can be combined for comprehensive analysis [13].
The ATR inhibitor study demonstrated the value of parallel screening approaches. Researchers performed both CRISPR knockout screens using the Brunello library (targeting 19,114 genes with 76,441 gRNAs) and CRISPR activation screens to identify genes regulating resistance to VE822 and AZD6738 inhibitors [13]. This dual approach revealed that different genes and pathways confer resistance when knocked out versus when overexpressed, providing a more complete understanding of the resistance landscape.
Practical Implementation and Workflow
The experimental workflow for dual CRISPR screens involves several critical steps. First, cells are infected with the CRISPR library at sufficient coverage (e.g., 20 million cells for 250-fold library coverage) [13]. The population is then divided into treatment and control groups, with treatment groups exposed to selective pressure (e.g., ATR inhibitors) while controls receive vehicle alone [13]. After a sufficient selection period, genomic DNA is extracted from surviving cells, gRNA sequences are amplified and sequenced, and bioinformatic analysis identifies enriched or depleted gRNAs [13].
The IMPACT computational framework enhances screen interpretation by integrating multi-parametric phenotypic data with protein interaction networks [105]. This approach helps identify consistent phenotypic patterns among interacting genes, rescues genes with weak phenotypes, and accounts for multiple biases in screen data [105]. The method applies either gene set-based analysis (IMPACT-sets) to find enriched phenotypes in pre-defined gene groups, or network-based analysis (IMPACT-modules) to identify sub-networks with common phenotypic signatures [105].
Diagram 1: Workflow for dual CRISPR screening approaches. The diagram illustrates the parallel implementation of knockdown and activation screens under different selection pressures, leading to identification of both resistance and sensitivity phenotypes, followed by integrated computational analysis.
Key Signaling Pathways and Molecular Mechanisms
Pathway-Level Insights from Opposing Manipulations
The molecular mechanisms underlying resistance and sensitivity phenotypes often involve well-defined signaling pathways. In the P2X7 receptor study, researchers demonstrated that both overexpression and knockdown experiments affected the PI3K/Akt/GSK-3β pathway, JNK signaling, and epithelial-mesenchymal transition (EMT) [27]. This pattern suggests these pathways as central mechanisms through which P2X7 receptor expression influences cancer progression.
The cyclin D1 and p27Kip1 study in Müller glia cells provides another compelling example of how opposing manipulations can reveal regulatory mechanisms. Here, simultaneous cyclin D1 overexpression and p27Kip1 knockdown synergistically promoted robust cell cycle re-entry in normally quiescent cells [96]. Neither manipulation alone achieved the same effect, demonstrating how complementary approaches can reveal functional interactions between pathway components. Single-cell RNA sequencing further showed that cell cycle reactivation led to suppression of interferon signaling, activation of reactive gliosis, and downregulation of glial genes [96].
Network Biology and Complex Interactions
Beyond linear pathways, opposing phenotypes often emerge from complex network interactions. Dosage chromosome instability (dCIN) genes identified through overexpression screens exemplify this principle [50]. Overexpression of 245 yeast genes caused chromosome instability, with human orthologs frequently overexpressed or amplified in tumors [50]. This discovery enabled identification of synthetic dosage lethal (SDL) interactions, where overexpression of one gene creates vulnerability to inhibition of an unrelated gene [50]. For example, TDP1 overexpression created sensitivity to histone deacetylase inhibitors, revealing a therapeutically exploitable vulnerability [50].
Diagram 2: Molecular pathways underlying opposing phenotypes. The diagram illustrates how manipulation of the P2X7 receptor affects multiple signaling pathways, leading to contrasting cellular phenotypes, while synergistic effects emerge from combined cyclin D1 overexpression and p27Kip1 knockdown.
Research Reagent Solutions for Screening Experiments
Successful resistance and sensitivity screens require carefully selected reagents and methodologies. The table below summarizes key solutions for implementing these experiments.
Table 2: Essential Research Reagents for Resistance and Sensitivity Screens
| Reagent/Library | Type | Primary Application | Key Features |
|---|---|---|---|
| Brunello CRISPR Knockout Library | CRISPRko | Genome-wide knockout screens | Targets 19,114 genes with 76,441 gRNAs; ~4 gRNAs/gene [13] |
| Calabrese CRISPR Activation Library | CRISPRa | Genome-wide overexpression screens | Enables systematic gene overexpression studies [13] |
| AAV Vectors (e.g., AAV7m8) | Viral Delivery | In vivo and in vitro gene delivery | Cell-type specific expression (e.g., GFAP promoter for Müller glia) [96] |
| IMPACT Framework | Computational Tool | Multi-parametric data integration | Combines phenotypic profiles with network information; reduces noise [105] |
| RSA Algorithm | Bioinformatics | Hit identification from screen data | Ranks genes based on gRNA enrichment/depletion patterns [13] |
Experimental Design Considerations
Selecting appropriate screening conditions requires careful consideration of multiple factors. For resistance screens, optimal detection requires high drug pressure (70-90% growth inhibition) to create strong selective pressure for resistant clones [104]. Sensitivity screens perform better with moderate drug pressure (10-30% growth inhibition) to allow detection of enhanced vulnerability without complete cell death [104]. The ATR inhibitor study used 1.5μM VE822 and 3.6μM AZD6738, concentrations that kill approximately 90% of cells over 108 hours, creating ideal conditions for resistance detection [13].
Library selection significantly impacts screen quality, particularly for sensitivity screens that benefit from higher numbers of guides per gene to provide statistical robustness for detecting dropout phenotypes [104]. For both screening types, maintaining sufficient library coverage (typically 250-500x) ensures faithful representation of all perturbations throughout the experiment [13].
Resistance and sensitivity phenotypes provide complementary insights into gene function and biological mechanisms. Resistance screens identify genes whose perturbation confers survival advantages under selective pressure, while sensitivity screens reveal vulnerabilities that enhance compound effects. The strategic application of both knockdown and overexpression approaches, combined with appropriate selective pressures and computational analysis methods, enables comprehensive understanding of gene function and network biology. As screening technologies advance, integrating these opposing phenotypic perspectives will continue to drive discoveries in basic biology and therapeutic development, particularly through multi-parametric assays and network-based analysis frameworks like IMPACT that capture the complexity of biological systems.
In the modern drug development pipeline, genetic screens are indispensable for bridging the gap between basic cellular biology and viable therapeutic targets. Two powerful, complementary approaches—overexpression and knockdown/knockout screens—enable researchers to systematically identify genes that influence disease processes. Overexpression screens investigate the consequences of increasing a gene's expression level, mimicking oncogenic activation or therapeutic protein supplementation. In contrast, knockdown/knockout screens assess what happens when gene function is partially or completely abolished, simulating therapeutic inhibition [83] [23].
The choice between these approaches is not merely technical but philosophical, influencing which types of drug targets a campaign will prioritize. Knockout screens traditionally excel at identifying genes whose function is essential for a disease process, pointing to inhibitory targets like small molecules or therapeutic antibodies. Overexpression screens naturally reveal genes with sufficient activity to suppress a pathological state, suggesting opportunities for protein replacement therapy or agonist development. This guide provides a comparative analysis of these methodologies to inform strategic decision-making in therapeutic development.
Theoretical Foundations: Mechanisms and Consequences
Quantitative Principles of Overexpression
A critical distinction often overlooked is that overexpression experiments are inherently quantitative rather than qualitative. The cellular consequences depend not just on which gene is overexpressed, but to what degree. Two primary experimental paradigms exist:
- Absolute Overexpression: Achieved through strong constitutive promoters (e.g., GAL promoter in yeast) on multi-copy plasmids, this method drives target proteins to similarly high concentrations regardless of their native expression levels. This approach creates massive overexpression folds for normally low-abundance proteins and more modest folds for already abundant ones [83].
- Relative Overexpression: Accomplished by increasing gene copy number using vectors maintained at a specific copy number, this method increases expression relative to the native level. This results in higher absolute protein levels for genes that are already highly expressed natively [83].
The mechanistic consequences of protein overexpression that can cause cellular defects include:
- Resource Overload: Squandering cellular energy and metabolic precursors on unnecessary protein synthesis.
- Stoichiometric Imbalance: Disrupting multi-protein complexes by flooding them with one subunit.
- Promiscuous Interactions: Enabling low-affinity, non-physiological binding that would not occur at normal concentrations.
- Pathway Modulation: Artificially activating or suppressing signaling cascades [83].
Principles of Gene Knockdown and Knockout
Knockdown and knockout represent a graded series of techniques for reducing gene function:
- Gene Knockdown: Typically achieved via RNA interference (RNAi) using siRNA or shRNA, this technique partially reduces gene expression by targeting mRNA for degradation, producing a hypomorphic phenotype that allows study of essential genes [23] [106].
- Gene Knockout: Completely abolishes gene function through methods like CRISPR-Cas9, which introduces frameshift mutations in the coding sequence, or homologous recombination in embryonic stem cells [48] [23].
Each method has distinct strengths for therapeutic prioritization. Knockdown is invaluable for studying essential genes whose complete loss would be lethal, while knockout provides unambiguous evidence of a gene's function and can reveal compensatory mechanisms that evolve in its absence.
Methodological Comparison: Experimental Designs and Workflows
Core Experimental Protocols
Genome-Wide CRISPR Knockout Screen Protocol [13] [48]
- Library Design: Utilize a genome-wide sgRNA library (e.g., Brunello library with 76,441 gRNAs targeting 19,114 genes) [13].
- Cell Engineering: Stably express Cas9 nuclease in the target cell line (e.g., J-Lat 10.6_Cas9 for HIV latency studies) [48].
- Library Transduction: Transduce cells with the sgRNA library at low MOI to ensure single integration events.
- Selection: Apply puromycin selection for 7-21 days to eliminate non-transduced cells.
- Experimental Arm: Apply the selective pressure (e.g., ATR inhibitors VE822 or AZD6738; monitoring HIV-1 reactivation via GFP) [13] [48].
- Control Arm: Maintain cells under standard conditions.
- Genomic DNA Extraction: Harvest genomic DNA from both populations after selection period.
- sgRNA Amplification & Sequencing: PCR-amplify sgRNA cassettes for Illumina sequencing.
- Bioinformatic Analysis: Use algorithms like MAGeCK or RSA to identify significantly enriched or depleted sgRNAs, indicating genes conferring resistance or sensitivity [13] [48].
CRISPR Activation (Overexpression) Screen Protocol [13]
- Library Design: Employ an activation library (e.g., Calabrese library) with sgRNAs targeting transcriptional start sites or activating elements.
- Cell Engineering: Stably express a catalytic dead Cas9 (dCas9) fused to transcriptional activators like VP64-p65-Rta (VPR).
- Library Transduction: Transduce at low MOI as in knockout screens.
- Selection & Analysis: Follow similar selection, sequencing, and analysis steps as knockout screens to identify genes whose overexpression confers a selective advantage [13].
Focused cDNA Overexpression Screen Protocol [28] [32]
- Library Design: Curate a targeted cDNA library (e.g., 163 chromosome 21 genes or 414 interferon-stimulated genes) cloned into lentiviral vectors [28] [32].
- Arrayed Format: Often conducted in "one gene per well" format rather than pooled approach.
- Transduction: Individually transduce each cDNA construct into target cells.
- Phenotypic Assessment: Apply relevant assay (e.g., Sonic Hedgehog signaling response or Toxoplasma gondii infection resistance) [28] [32].
- Hit Identification: Identify cDNAs that significantly modulate the measured phenotype compared to controls.
Workflow Visualization
The following diagram illustrates the parallel workflows for knockout and activation screens, highlighting their shared steps and key differences:
Diagram 1: Parallel workflows for knockout and overexpression screens.
Comparative Analysis: Strategic Implications for Target Prioritization
Direct Methodology Comparison
Table 1: Strategic comparison of screening approaches for therapeutic target identification
| Parameter | Knockdown/Knockout Screens | Overexpression Screens |
|---|---|---|
| Therapeutic Analogy | Inhibitory drugs (small molecules, therapeutic antibodies) | Protein replacement therapy, agonist drugs |
| Primary Mechanism | Loss-of-function (complete or partial) | Gain-of-function |
| Gene Types Identified | Essential genes, disease promoters | Tumor suppressors, pathogenic suppressors |
| Typical Hit Rate | Relatively higher | Relatively lower |
| False Positives | Off-target effects (RNAi), genetic compensation | Non-physiological expression levels, promiscuous interactions |
| False Negatives | Redundant genes, essential genes | Genes requiring post-translational modification |
| Key Applications | Identifying essential host factors for pathogens [48], synthetic lethal interactions [13] | Identifying restriction factors for pathogens [32], modulators of signaling pathways [28] |
| Therapeutic Development | Directly identifies inhibitor targets | Directly identifies protein therapy targets & agonist targets |
Quantitative Outcomes from Comparative Studies
Table 2: Representative quantitative outcomes from published genetic screens
| Study Objective | Screening Approach | Key Quantitative Findings | Therapeutic Implications |
|---|---|---|---|
| Identify ATR inhibitor resistance mechanisms [13] | Dual genome-wide CRISPR knockout & activation | 393 genes (VE822) and 456 genes (AZD6738) conferred resistance when knocked out; 118 genes overlapped both screens | Identified MED12-TGFβ pathway as regulating replication fork stability; potential biomarkers for ATR inhibitor response |
| Discover HIV-1 latency regulators [48] | Genome-wide CRISPR knockout in J-Lat 10.6 cells | 211 significantly enriched genes (P<0.01); identified PSMD1, UCH37, USP14 as latency regulators | Deubiquitinases represent novel targets for latency reversal agents ("shock and kill" strategy) |
| Find ISGs restricting Toxoplasma gondii [32] | cDNA overexpression (414 ISGs) | Identified RARRES3 as a potent restrictor; limited number of individual ISGs showed restrictive activity | Suggests IFNγ-mediated immunity is multifactorial; RARRES3 represents a novel host-directed therapy target |
| Identify SHH pathway modulators in Down syndrome [28] | Chromosome 21 cDNA overexpression (163 genes) | DYRK1A upregulated, while B3GALT5, ETS2, HMGN1, MIS18A inhibited SHH signaling | Prioritizes dosage-sensitive targets for ameliorating Down syndrome phenotypes |
Mechanistic Insights Visualization
The cellular outcomes from screening hits follow predictable mechanistic patterns that directly inform therapeutic strategy:
Diagram 2: Mechanistic outcomes and corresponding therapeutic strategies.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key research reagents for implementing genetic screens
| Reagent Category | Specific Examples | Function & Application |
|---|---|---|
| CRISPR Knockout Libraries | Brunello library (76,441 gRNAs) [13], GeCKO v2 library (123,411 gRNAs) [48] | Genome-wide loss-of-function screening; 4-6 gRNAs/gene improves statistical confidence |
| CRISPR Activation Libraries | Calabrese library [13] | Genome-wide gain-of-function screening using dCas9-activator fusions |
| cDNA Overexpression Libraries | Chromosome 21 cDNA library (163 genes) [28], ISG library (414 genes) [32] | Targeted overexpression screening in arrayed format; identifies suppressors |
| Validation Reagents | siRNA/shRNA [24], Individual gRNAs [48], Pharmacological inhibitors [48] [24] | Confirm screening hits through orthogonal approaches |
| Analytical Tools | MAGeCK [48], RSA algorithm [13] | Statistical identification of significantly enriched/depleted genes |
| Detection Methods | Quantitative Western blotting, In-Cell Western Assay [82] | Confirm knockout/knockdown efficiency at protein level |
Integrated Prioritization Framework: From Screen to Drug Target
Successfully translating screening hits into viable therapeutic programs requires a systematic triage process:
Technical Validation: Confirm phenotypes using orthogonal methods (e.g., validate CRISPR hit with siRNA or pharmacological inhibitor) [48] [24]. Assess protein-level changes via Western blotting [82].
Therapeutic Triage: Categorize hits based on druggability and therapeutic strategy. Knockout hits suggesting essential disease genes are natural small molecule inhibitor targets. Overexpression hits suggesting disease suppression may indicate protein therapeutic or gene therapy opportunities.
Mechanistic Deconvolution: Elucidate the precise molecular mechanism. For example, the discovery that MED12 regulates replication fork stability via TGFβ signaling provides a mechanistic basis for ATR inhibitor resistance [13].
Pathway Contextualization: Position hits within broader signaling networks. The identification of multiple deubiquitinases (UCH37, USP14, OTULIN) in HIV-1 latency suggests a central role for ubiquitination in viral persistence [48].
Biomarker Potential: Evaluate hits as potential predictive biomarkers. Genes conferring resistance to ATR inhibitors could stratify patients most likely to respond to therapy [13].
The most powerful screening strategies often combine both approaches, as demonstrated in the dual CRISPR knockout and activation screen for ATR inhibitor resistance [13]. This comprehensive mapping of the genetic landscape surrounding a therapeutic target provides the most robust foundation for therapeutic development, identifying both resistance mechanisms and sensitizing factors within a single experimental framework.
Conclusion
Overexpression and knockdown screens are not mutually exclusive but are powerfully complementary tools in the functional genomics arsenal. Overexpression excels at identifying sufficient genes, drug resistance mechanisms, and potential therapeutic targets, while knockdown screens are unparalleled for finding essential genes, vulnerabilities, and validating target engagement. The advent of precise CRISPRa and CRISPRi systems has mitigated many historical limitations, enabling more reliable and interpretable results. The future lies in the intelligent integration of both approaches, as demonstrated by dual screening strategies that provide a more complete mechanistic understanding of complex biological processes. For drug discovery, this combined path accelerates the identification of novel targets, the understanding of drug mode of action, and the prediction of resistance, ultimately paving the way for more personalized and effective cancer therapies and treatments for neurodegenerative disorders. Future directions will focus on improving predictive in silico models, expanding into more physiologically relevant primary cell models, and leveraging single-cell multi-omics readouts to deconvolute complex cellular responses to genetic perturbation.
References
- 1. Prevalence of loss-of-function, gain-of-function and ... [https://www.nature.com/articles/s41467-025-63234-3]
- 2. Overexpression of KDM6A in Hepatoma Cells Induces ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12619137/]
- 3. Comparative analysis of antibody-mediated loss ... [https://www.sciencedirect.com/science/article/pii/S2472555225000760]
- 4. KAT6B overexpression in mice causes aggression, anxiety ... [https://www.sciencedirect.com/science/article/pii/S2589004225002135]
- 5. TDP-43 loss induces cryptic polyadenylation in ALS/FTD - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12586162/]
- 6. A comparison of computational methods for expression ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03840-y]
- 7. Simultaneous cyclin D1 overexpression and p27kip1 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11968108/]
- 8. Lambert-GA | CRISPR-interference [https://2025.igem.wiki/lambert-ga/therapeutics-crispri]
- 9. CRISPR interference [https://horizondiscovery.com/en/applications/crisprmod/crispri]
- 10. Positive correlations between knockdown and overexpression profiles from LINCS L1000 | Thinklab [https://think-lab.github.io/d/171/]
- 11. CRISPR Statistics and Facts (2025) [https://media.market.us/crispr-statistics/]
- 12. Addgene: Pooled Libraries [https://www.addgene.org/pooled-library/]
- 13. Dual genome-wide CRISPR knockout and CRISPR activation screens identify mechanisms that regulate the resistance to multiple ATR inhibitors | PLOS Genetics [https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009176]
- 14. A benchmark comparison of CRISPRn guide-RNA design algorithms... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11863645/]
- 15. /Cas-based customization of pooled CRISPR CRISPR libraries [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0199473]
- 16. AI-powered CRISPR could lead to faster gene therapies, Stanford Medicine study finds [https://med.stanford.edu/news/all-news/2025/09/ai-crispr-gene-therapy.html]
- 17. Engineering novel CRISPRi repressors for highly efficient mammalian gene regulation | Genome Biology | Full Text [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03640-4]
- 18. CRISPR-Cas-mediated transcriptional modulation: The therapeutic... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10362391/]
- 19. and dCas9-VPR reagents for CRISPR activation... CRISPRa guide RNA [https://horizondiscovery.com/en/crisprmod/crispra]
- 20. CRISPR-based Activation of Endogenous Neurotrophic Genes in... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6735509/]
- 21. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6093183/]
- 22. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5886776/]
- 23. Gene Knockdown - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/medicine-and-dentistry/gene-knockdown]
- 24. Overexpression and knock-down studies highlight that a disintegrin and metalloproteinase 28 controls proliferation and migration in human prostate cancer - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5059087/]
- 25. Gene knockdown - Wikipedia [https://en.wikipedia.org/wiki/Gene_knockdown]
- 26. Gene Knockdown - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/gene-knockdown]
- 27. or Overexpression of the P2X7 receptor regulates the... knockdown [https://pubmed.ncbi.nlm.nih.gov/39993700/]
- 28. of chromosome 21 genes reveals modulators... Overexpression screen [https://pmc.ncbi.nlm.nih.gov/articles/PMC10120076/]
- 29. RNA Interference to Knock Down Gene Expression - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6743327/]
- 30. Frontiers | Double UP: A Dual Color, Internally Controlled Platform for... [https://www.frontiersin.org/journals/molecular-neuroscience/articles/10.3389/fnmol.2020.00082/full]
- 31. Frontiers | Construction of EpCAM Overexpression and Knockdown ... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2025.1679698/abstract]
- 32. of interferon-stimulated genes identifies... Overexpression screen [https://elifesciences.org/articles/73137]
- 33. Rapid and robust validation of pooled CRISPR knockout screens using CelFi | Scientific Reports [https://www.nature.com/articles/s41598-025-96095-3]
- 34. Genome-scale overexpression reveals membrane... screen [https://pubmed.ncbi.nlm.nih.gov/40651551/]
- 35. Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs | Nature Communications [https://www.nature.com/articles/s41467-023-38119-y]
- 36. RNA interference as a key to knockdown overexpressed cyclooxygenase-2 gene in tumour cells | British Journal of Cancer [https://www.nature.com/articles/6603094]
- 37. CRISPRa and CRISPRi - Synthego [https://www.synthego.com/guide/crispr-methods/crispri-crispra/]
- 38. CRISPRa and CRISPRi: Why You Need These Powerful Tools [https://bitesizebio.com/47575/crispra-and-crispri/]
- 39. Compact and highly active next-generation libraries for... [https://elifesciences.org/articles/19760]
- 40. Optimized libraries for CRISPR-Cas9 genetic... | Nature Communications [https://www.nature.com/articles/s41467-018-07901-8?error=cookies_not_supported]
- 41. Frontiers | CRISPR activation: identifying and using novel genes for... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2025.1596600/full]
- 42. A CRISPRi /a platform in human iPSC-derived... | Nature Neuroscience [https://www.nature.com/articles/s41593-022-01131-4?error=cookies_not_supported&code=89cf8931-f1c9-4cc3-8ba7-9c6a24e3f44e]
- 43. Genome-wide CRISPRi /a screens in human neurons link lysosomal... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8254803/]
- 44. Systematic comparison of CRISPR-Cas9 and RNAi screens for essential genes - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4900911/]
- 45. Surfaceome CRISPR screen identifies OLFML3 as... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-021-02513-w]
- 46. A genome-scale screen for synthetic drivers of T-cell proliferation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9908437/]
- 47. Dual genome-wide CRISPR knockout and CRISPR activation screens ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7660927/]
- 48. CRISPR-based gene knockout screens reveal deubiquitinases involved in HIV-1 latency in two Jurkat cell models | Scientific Reports [https://www.nature.com/articles/s41598-020-62375-3]
- 49. Are Useful Cancer for Medical Research... Cell Lines Model Systems [https://pmc.ncbi.nlm.nih.gov/articles/PMC6721418/]
- 50. Overexpression screens identify conserved dosage chromosome instability genes in yeast and human cancer - PubMed [https://pubmed.ncbi.nlm.nih.gov/27551064/]
- 51. for Cell medicine in line (Review) modeling systems cancers [https://www.spandidos-publications.com/10.3892/ijo.2013.2202?text=fulltext]
- 52. Using induced pluripotent stem cells derived neurons to model brain diseases - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5558480/]
- 53. How to differentiate induced pluripotent stem cells into sensory neurons for disease modelling: a functional assessment | Stem Cell Research & Therapy | Full Text [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-03696-2]
- 54. Microglia heterogeneity, modeling and cell-state annotation in development and neurodegeneration | Nature Neuroscience [https://www.nature.com/articles/s41593-025-01931-4]
- 55. Evolving Models and Tools for Microglial Studies in the Central Nervous System - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8353053/]
- 56. ATR Inhibitor: Mechanism, Classes, and Research Trends - Biology Insights [https://biologyinsights.com/atr-inhibitor-mechanism-classes-and-research-trends/]
- 57. Genome-wide CRISPR /Cas9 library screening identified ATM... [https://ehoonline.biomedcentral.com/articles/10.1186/s40164-023-00416-z]
- 58. Bioinformatics approaches to analyzing CRISPR screen data: from dropout screens to single-cell CRISPR screens - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10019185/]
- 59. A benchmark of algorithms for the analysis of pooled CRISPR screens | Genome Biology | Full Text [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-01972-x]
- 60. Towards elucidating disease-relevant states of neurons and glia by... [https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-022-01134-7]
- 61. in Drug Discovery: An... | Technology Networks Phenotypic Screening [https://www.technologynetworks.com/drug-discovery/articles/phenotypic-screening-a-powerful-tool-for-drug-discovery-398572]
- 62. CRISPR-based functional genomics screening in... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10203043/]
- 63. The power of sophisticated phenotypic screening and modern mechanism-of-action methods - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4779180/]
- 64. Automating optical pooled screening [https://www.news-medical.net/health/Automating-optical-pooled-screening-with-massively-multiplexed-phenotypic-readouts.aspx]
- 65. M-CSF increases proliferation and phagocytosis while modulating... [https://jneuroinflammation.biomedcentral.com/articles/10.1186/1742-2094-10-85]
- 66. Multiple Genetic Interaction Experiments Provide Complementary... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002559]
- 67. Maximizing CRISPRi efficacy and accessibility with dual - sgRNA ... [https://elifesciences.org/articles/81856]
- 68. Genome-wide CRISPR -Cas9 knockout screens - Wikipedia [https://en.wikipedia.org/wiki/Genome-wide_CRISPR-Cas9_knockout_screens]
- 69. Complementary CRISPR genome-wide genetic screens in... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9518689/]
- 70. Arrayed CRISPR libraries for the... | Nature Biomedical Engineering [https://www.nature.com/articles/s41551-024-01278-4?error=cookies_not_supported&code=5ebef475-8568-4d66-919d-698ee5232165]
- 71. Integration of Organoids With CRISPR Screens: A Narrative Review - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11995251/]
- 72. Methods and applications of in vivo CRISPR screening | Nature Reviews Genetics [https://www.nature.com/articles/s41576-025-00873-8]
- 73. Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4922510/]
- 74. CRISPR Screen Explained: Methods, Applications & Design | Ubigene [https://www.ubigene.us/application/crispr-library.html]
- 75. CRISPR Activators: A Comparison Between dCas -VP64, SAM... 9 [https://blog.addgene.org/crispr-activators-dcas9-vp64-sam-suntag-vpr]
- 76. Team:Freiburg/Project/method - 2013.igem.org [https://2013.igem.org/Team:Freiburg/Project/method]
- 77. Genes adapt to outsmart gene-targeting strategies in mutant mouse strains by skipping exons to reinitiate transcription and translation | Genome Biology | Full Text [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-02086-0]
- 78. Diverse Roles of the Exon Junction Complex Factors in the Cell Cycle... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9499366/]
- 79. Combined shRNA over CRISPR/cas9 as a methodology to detect off-target effects and a potential compensatory mechanism | Scientific Reports [https://www.nature.com/articles/s41598-017-18551-z]
- 80. Cas13d-mediated isoform-specific RNA knockdown with a unified... [https://thenote.app/post/en/cas13d-mediated-isoform-specific-rna-knockdown-with-a-unified-computational-and-rmtlpbpntg]
- 81. and applications of Construction -trapping gene- exon ... targeting [https://bmcresnotes.biomedcentral.com/articles/10.1186/s13104-015-1241-6]
- 82. Confirm Target Gene Knockdown or Knockout with Fluorescent RNAi Studies [https://www.licorbio.com/applications/gene-knockdown-knockout]
- 83. nature of Quantitative - PMC overexpression experiments [https://pmc.ncbi.nlm.nih.gov/articles/PMC4710226/]
- 84. Improved pathway reconstruction from RNA interference screens by... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6022657/]
- 85. A genome scale overexpression screen to reveal drug activity in human cells | Genome Medicine | Full Text [https://genomemedicine.biomedcentral.com/articles/10.1186/gm549]
- 86. Diversity by Design: Addressing Mode Collapse Improves scRNA-seq... [https://arxiv.org/html/2506.22641v1]
- 87. Perturb-tracing enables high-content screening of multi-scale 3D genome regulators | Nature Methods [https://www.nature.com/articles/s41592-025-02652-z]
- 88. Massively parallel in vivo Perturb-seq screening - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12237601/]
- 89. A pragmatic approach to hit validation following biochemical high-throughput screening [https://www.drugtargetreview.com/article/27913/hit-validation-high-throughput-screening/]
- 90. A CRISPRi/a platform in human iPSC-derived microglia uncovers... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9448678/]
- 91. High-Throughput Screening | Assay.Works [https://www.assay.works/services/high-throughput-screening]
- 92. SciLifeLab & KNIME Help Labs Asses Raw Data from HTS | KNIME [https://www.knime.com/blog/a-workflow-for-high-throughput-screening-data-analysis-processing-and-hit-identification]
- 93. Simultaneous cyclin D1 overexpression and p27kip1 knockdown ... [https://elifesciences.org/reviewed-preprints/100904]
- 94. Genome-scale CRISPR-Cas9 Knockout and Transcriptional Activation Screening - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5526071/]
- 95. CRISPR approaches to small molecule target identification - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5834945/]
- 96. Simultaneous cyclin D1 overexpression and p27kip1 knockdown ... [https://elifesciences.org/articles/100904]
- 97. Peer review in Simultaneous cyclin D1 overexpression and... [https://elifesciences.org/articles/100904/peer-reviews]
- 98. A High-Throughput Platform for Lentiviral Overexpression Screening of the Human ORFeome | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0020057]
- 99. Synergistic drug combinations improve therapeutic selectivity - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2708317/]
- 100. Integrating Multiple Correlated Phenotypes for Genetic Association Analysis by Maximizing Heritability - PubMed [https://pubmed.ncbi.nlm.nih.gov/26111731/]
- 101. Increase your recognition in the scientific world with short video-casts [https://sciencecast.org/casts/x2f7aylgncwi]
- 102. of cell cycle regulator CDCA3 promotes oral cancer... Overexpression [https://bmccancer.biomedcentral.com/articles/10.1186/1471-2407-12-321]
- 103. predicts the prognosis of advanced serous ovarian cancer Cyclin D 1 [https://www.spandidos-publications.com/10.3892/etm.2011.194]
- 104. CRISPR screening strategies: resistance vs. sensitivity - what’s right for your study? | Revvity [https://www.revvity.com/blog/crispr-screening-strategies-resistance-vs-sensitivity-whats-right-your-study]
- 105. Revealing Molecular Mechanisms by Integrating High-Dimensional... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4154648/]
- 106. RNA Knockdown - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/immunology-and-microbiology/rna-knockdown]