Automating High-Throughput NGS for Chemogenomics: Strategies for Scalable Drug Discovery
This article provides a comprehensive guide for researchers and drug development professionals on implementing automated Next-Generation Sequencing (NGS) workflows for high-throughput chemogenomics.
Automating High-Throughput NGS for Chemogenomics: Strategies for Scalable Drug Discovery
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on implementing automated Next-Generation Sequencing (NGS) workflows for high-throughput chemogenomics. It explores the foundational drivers of automation, including the need for scalability and reproducibility in large-scale drug screening. The scope covers practical methodologies for integrating liquid handling systems and end-to-end platforms, strategies for overcoming common bottlenecks in data analysis and library preparation, and frameworks for the rigorous validation required in regulated research environments. By synthesizing current technological advancements with practical application, this resource aims to equip scientists with the knowledge to accelerate target identification and therapeutic discovery.
The Rise of Automation in NGS: Fueling the Next Generation of Chemogenomics
The global landscape of genomic analysis is undergoing a rapid transformation, driven by an unprecedented convergence of technological advancement, declining costs, and expanding applications across biomedical research and clinical diagnostics. The genome reconstruction tools market, a specialized segment of the bioinformatics industry, is projected to grow from USD 182.6 million in 2025 to USD 387 million by 2035, reflecting a compound annual growth rate (CAGR) of 7.8% [1]. This growth is fundamentally fueled by the critical need for scalable analytical solutions that can manage the enormous data volumes generated by modern sequencing technologies. Concurrently, the broader genome testing market demonstrates even more accelerated expansion, expected to rise from USD 22.45 billion in 2025 to USD 55.23 billion by 2032, at a CAGR of 13.7% [2]. These markets are being reshaped by the pervasive adoption of cloud-based bioinformatics platforms across biotechnology and pharmaceutical sectors, alongside a pronounced shift toward precision medicine tools in both research and clinical applications [1].
For researchers and drug development professionals, this expansion creates both opportunities and challenges. The ability to process and interpret massive genomic datasets efficiently has become a critical bottleneck in high-throughput chemogenomics research, where the rapid profiling of chemical-genetic interactions is essential for target identification and validation. This application note examines the key drivers behind this growing demand for scalability and provides detailed protocols for implementing automated, high-throughput genomic workflows that address these challenges directly.
Table 1: Key Market Growth Indicators for Genomic Analysis Technologies
| Market Segment | 2025 Projected Value | 2035/2032 Projected Value | CAGR | Primary Growth Drivers |
|---|---|---|---|---|
| Genome Reconstruction Tools | USD 182.6 million [1] | USD 387 million [1] | 7.8% [1] | Cloud/SaaS adoption, pharmaceutical R&D, microorganism analysis [1] |
| Whole Genome Sequencing | USD 3 billion [3] | USD 6.1 billion (2030) [3] | 15.1% [3] | Cancer genomics, rare disease research, personalized medicine [3] |
| Functional Genomics | USD 11.34 billion [4] | USD 28.55 billion (2032) [4] | 14.1% [4] | NGS technology advances, drug discovery applications [4] |
| Genome Testing | USD 22.45 billion [2] | USD 55.23 billion (2032) [2] | 13.7% [2] | Clinical diagnostics, direct-to-consumer testing, pharmacogenomics [2] |
Key Market Drivers for Scalable Genomic Analysis
Technological Advancements and Cost Reduction
The dramatic reduction in sequencing costs has been the most fundamental driver accelerating demand for scalable genomic analysis. Since the completion of the Human Genome Project, the cost of sequencing a full human genome has decreased by approximately 96% [5], making large-scale genomic studies economically feasible for more laboratories. This cost reduction has been coupled with substantial improvements in sequencing throughput and capabilities. Modern NGS technologies can simultaneously sequence millions to billions of DNA fragments in a massively parallel fashion, enabling researchers to expand the scale and discovery power of their genomic studies far beyond what was possible with traditional techniques [5].
Leading instrument companies are continuously pushing the boundaries of sequencing performance. For instance, Ultima Genomics' UG 100 Solaris system, launched in 2025, offers a >50% increase in output to 10-12 billion reads per wafer while reducing the price to $0.24 per million reads, potentially enabling the $80 genome [6]. Similarly, Roche's introduction of Sequencing by Expansion (SBX) technology represents a significant innovation that uses biochemical conversion to encode DNA into surrogate Xpandomer molecules, enabling highly accurate single-molecule nanopore sequencing [6]. These technological advancements are critically important for chemogenomics research, where profiling thousands of chemical compounds across multiple cell lines requires unprecedented sequencing scale and cost-efficiency.
Expanding Applications in Biomedical Research and Clinical Diagnostics
The applications driving demand for scalable genomic analysis span virtually all areas of biomedical research and are increasingly penetrating clinical diagnostics. In cancer genomics and rare inherited diseases, whole genome sequencing has become indispensable for identifying genetic mutations and enabling faster, more accurate diagnoses [3]. The growing focus on targeted therapies and personalized medicine further fuels this demand, as WGS supports treatment personalization by revealing genetic profiles that guide therapeutic decisions [3].
The functional genomics market, where NGS commands a dominant 32.5% technology share [4], exemplifies the broadening applications. Transcriptomics alone accounts for 23.4% of the functional genomics application segment [4], driven by expanding research on gene expression dynamics under different biological conditions. The microorganisms segment represents another substantial application area, accounting for 27.8% of genome reconstruction tools demand [1], with growing importance in microbiome research, infectious disease monitoring, and industrial biotechnology.
For drug development professionals, the integration of multi-omics approaches represents a particularly significant trend. The combination of genomic, proteomic, and metabolomic data provides unprecedented insights into disease mechanisms and therapeutic responses [2]. Additionally, pharmacogenomic testing services are expanding rapidly, enabling personalized medication management based on individual genetic profiles [2].
Adoption of Cloud Computing and Artificial Intelligence
The massive data volumes generated by modern genomic technologies have necessitated a fundamental shift in computational strategies. Cloud/SaaS subscription models have emerged as the leading service model in the genome reconstruction tools market, accounting for 32.8% market share [1]. These platforms provide the essential scalability and accessibility required for managing and analyzing large genomic datasets without substantial local computational infrastructure.
The integration of artificial intelligence and machine learning represents another transformative driver for scalable genomic analysis. As noted in the market research, "Efforts in standardization, artificial intelligence, and machine learning drive improvements in diagnostic reliability and processing speed, delivering value for both clinical and biopharmaceutical users" [2]. The development of sophisticated AI models specifically for genomic applications is accelerating, exemplified by initiatives such as the "Genos" AI model launched by BGI-Research and Zhejiang Lab in 2025 – the world's first deployable genomic foundation model with 10 billion parameters designed to analyze up to one million base pairs at single-base resolution [4].
Table 2: Key Technology Adoption Trends in Genomic Analysis
| Technology Trend | Market Impact | Application in Scalable Genomics |
|---|---|---|
| Cloud/SaaS Platforms | 32.8% market share in genome reconstruction tools [1] | Enables scalable data storage, computation, and collaboration for large-scale genomic studies |
| AI/ML Integration | Improving variant interpretation accuracy and processing speed [2] | Accelerates analysis of massive genomic datasets; enables pattern recognition in chemogenomic screens |
| Automation & High-throughput Workflows | Enables access to optimization space not possible using traditional laboratory work [7] | Increases sample processing capacity; reduces manual errors in library preparation |
| Multi-omics Approaches | Development of testing panels combining genome, proteome, and metabolome data [2] | Provides comprehensive view of biological systems for drug discovery and biomarker identification |
| NGS Technology | 32.5% share of functional genomics technology segment [4] | Foundation for high-throughput genomic analysis across diverse applications |
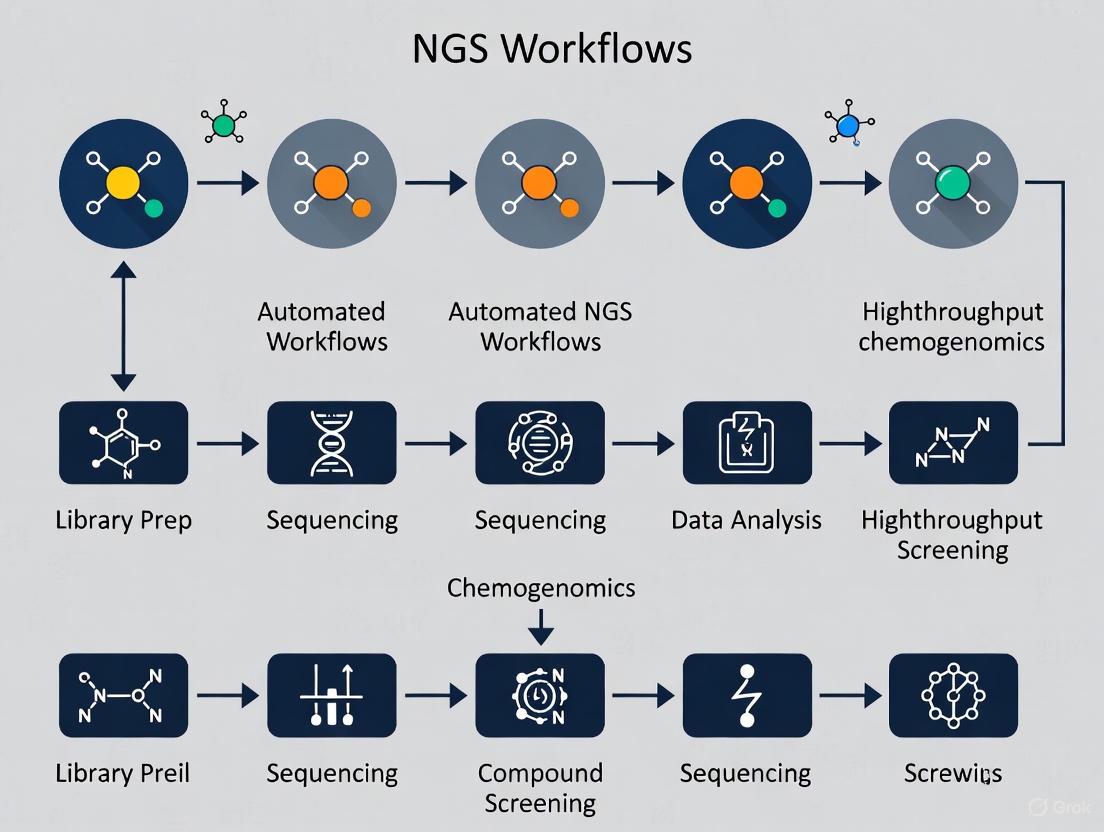
Automated NGS Workflow for High-Throughput Chemogenomics
The implementation of automated, high-throughput NGS workflows is particularly critical for chemogenomics research, which involves systematically profiling the interactions between chemical compounds and genomic elements. The optimization space for maximizing microbial conversions in biomanufacturing alone is vast, and "automation and rapid workflows can enable access to optimization space not possible using the throughput allowed by traditional laboratory work" [7]. For drug development professionals, this capability translates directly to accelerated target identification and validation cycles.
The fundamental NGS workflow comprises four key steps: nucleic acid extraction, library preparation, sequencing, and data analysis [5] [8]. In high-throughput chemogenomics applications, each of these steps presents specific scalability challenges that can be addressed through strategic automation and process optimization. The workflow detailed in this section has been specifically adapted for large-scale chemogenomic profiling, where processing hundreds to thousands of samples in parallel is essential for statistical power and discovery.
Protocol 1: Automated Nucleic Acid Extraction and Quality Control
Experimental Principle
The initial extraction phase is critical for generating high-quality sequencing data, particularly in chemogenomics applications where compound treatments may introduce inhibitors or affect nucleic acid integrity. Automated nucleic acid extraction ensures consistency across thousands of samples while minimizing cross-contamination risks – essential factors for reliable compound-genotype interaction studies [9].
Materials and Equipment
- Automated liquid handling system (e.g., Agilent Magnis NGS Prep System, Revvity chemagic 360)
- High-throughput nucleic acid extraction kits (compatible with automation)
- Cell lysis modules appropriate for your cell types
- Magnetic stand or plate handler for bead-based purification
- Real-time PCR system for quantification
- Fluorometric quantitation system (e.g., Qubit)
- Microfluidic electrophoresis system (e.g., Bioanalyzer, TapeStation)
Step-by-Step Procedure
Sample Plate Preparation: Arrange cell lysates from compound-treated samples in 96- or 384-well plates compatible with your automated liquid handling system. Include appropriate controls (untreated, vehicle controls, positive controls).
Automated Extraction Program:
- Program the liquid handling system to transfer 200μl of each lysate to a fresh processing plate.
- Add 20μl magnetic beads and 200μl binding buffer to each sample using automated pipetting.
- Incubate for 5 minutes at room temperature with periodic mixing.
- Engage magnetic stand for 2 minutes to separate beads from solution.
- Program aspiration of supernatant without disturbing bead pellets.
Wash Steps:
- Add 200μl wash buffer 1 to each well, resuspend beads, and incubate for 30 seconds.
- Engage magnetic stand, then aspirate supernatant.
- Repeat with wash buffer 2.
- Perform final wash with 80% ethanol.
Elution:
- Air-dry beads for 5-10 minutes to evaporate residual ethanol.
- Add 50μl elution buffer (10mM Tris-HCl, pH 8.5) to each well.
- Resuspend beads thoroughly and incubate for 2 minutes at room temperature.
- Engage magnetic stand and transfer 45μl of eluate to a fresh output plate.
Quality Control Assessment:
- Quantitate DNA/RNA yield using fluorometric methods (e.g., Qubit) with automated plate reading.
- Assess integrity via microfluidic electrophoresis (RIN >8.0 for RNA; distinct high molecular weight band for DNA).
- Verify purity through spectrophotometric ratios (A260/280: 1.8-2.0; A260/230: >2.0).
Critical Protocol Parameters
- Input Requirements: 10-1000ng DNA or 10-500ng RNA per sample
- Throughput: 96 samples in approximately 2 hours; 384 samples in 4 hours
- Success Indicators: Yield sufficient for library prep (>5ng/μl), high purity, minimal degradation
Protocol 2: Automated Library Preparation and Normalization
Experimental Principle
Automated library preparation standardizes the fragmentation, adapter ligation, and amplification steps required for NGS, eliminating the variability introduced by manual pipetting [9]. For chemogenomics applications, maintaining consistency across all samples is particularly crucial when comparing gene expression or mutation profiles across hundreds of compound treatments.
Materials and Equipment
- Automated library preparation kit (e.g., Illumina DNA Prep, KAPA HyperPrep)
- Dual-indexed adapter plates for sample multiplexing
- Automated liquid handling workstation with thermal cycling capability
- Magnetic separation module
- Real-time PCR quantification system
- Normalization and pooling automation
Step-by-Step Procedure
Fragmentation and End Repair:
- Program liquid handler to transfer 50-100ng of input DNA in 50μl volume to a PCR plate.
- Add 20μl fragmentation/end repair mix to each well.
- Seal plate and transfer to integrated thermal cycler: 5 minutes at 55°C, then hold at 4°C.
Adapter Ligation:
- Add 30μl ligation mix containing uniquely dual-indexed adapters to each well.
- Program addition of 15μl ligation enhancer and thorough mixing.
- Incubate for 15 minutes at 20°C using integrated temperature control.
Cleanup and Size Selection:
- Add 87μl bead-based cleanup solution to each well.
- Incubate 5 minutes at room temperature, engage magnets, and remove supernatant.
- Wash twice with 80% ethanol using automated dispensing and aspiration.
- Elute in 22μl resuspension buffer.
Library Amplification:
- Add 28μl PCR master mix to each well.
- Transfer to thermal cycler: 98°C for 45 seconds; [98°C for 15 seconds, 60°C for 30 seconds] × 8-12 cycles; 72°C for 1 minute.
- Perform final cleanup with 45μl beads, eluting in 25μl buffer.
Quality Control and Normalization:
- Quantify libraries using fluorometric methods with automated plate reading.
- Assess library size distribution via microfluidic electrophoresis (expected peak: 300-500bp).
- Program liquid handler to normalize all libraries to 4nM concentration based on quantification data.
- Combine equal volumes of normalized libraries into a sequencing pool.
Critical Protocol Parameters
- Input Requirements: 10-1000ng DNA or 10-500ng RNA per sample
- Throughput: 96 libraries in approximately 6 hours; 384 libraries in 9 hours
- Success Indicators: Appropriate library size distribution, concentration >2nM, minimal adapter dimer
Protocol 3: Automated Data Analysis Pipeline for Chemogenomics
Experimental Principle
The massive datasets generated in high-throughput chemogenomics require automated bioinformatic processing to extract meaningful biological insights. The analysis workflow progresses through three key stages: primary analysis (base calling, demultiplexing), secondary analysis (alignment, variant calling), and tertiary analysis (chemogenomic interpretation) [8]. Automation ensures consistency and enables the processing of hundreds of samples in parallel.
- High-performance computing cluster or cloud computing environment
- Workflow management system (e.g., Nextflow, Snakemake)
- Containerization platform (e.g., Docker, Singularity)
- Genomic analysis toolkit (e.g., GATK, Bioconductor)
- Custom scripts for chemogenomic profiling
Step-by-Step Procedure
Primary Analysis Setup:
- Configure workflow manager to process sequencing output from the instrument.
- Implement base calling and demultiplexing using bcl2fastq or similar tools.
- Program quality control checks (FastQC) with automated reporting.
Secondary Analysis Automation:
- Implement alignment to reference genome using BWA-MEM or STAR (for RNA-Seq).
- Program duplicate marking to remove PCR artifacts.
- Execute variant calling (HaplotypeCaller for DNA-Seq) or expression quantification (featureCounts for RNA-Seq).
Tertiary Analysis for Chemogenomics:
- Normalize expression counts or variant frequencies across all samples.
- Implement differential expression/analysis comparing each compound treatment to controls.
- Perform pathway enrichment analysis (GO, KEGG) for each compound.
- Generate compound-gene interaction networks.
Quality Monitoring and Reporting:
- Integrate automated quality metrics (alignment rate, duplication rate, coverage uniformity).
- Implement multiQC reporting for overall project assessment.
- Generate automated summary reports highlighting top compound-gene interactions.
Critical Protocol Parameters
- Computing Requirements: 32+ cores, 64GB+ RAM for typical datasets
- Processing Time: 4-24 hours depending on sample number and sequencing depth
- Success Indicators: High alignment rates (>90%), expected number of variants/expressed genes, identification of compound-specific signatures
Essential Research Reagent Solutions
The successful implementation of automated, high-throughput genomic workflows requires careful selection of reagents and materials specifically designed for automation compatibility and consistency. The following table details essential solutions for scalable chemogenomics research:
Table 3: Essential Research Reagent Solutions for Automated Genomic Workflows
| Reagent Category | Specific Product Examples | Function in Workflow | Automation Compatibility Features |
|---|---|---|---|
| Nucleic Acid Extraction Kits | Magnetic bead-based purification kits | Isolation of high-quality DNA/RNA from cell lysates | Pre-filled deep well plates, reduced incubation times, room temperature processing |
| Library Preparation Kits | Illumina DNA Prep, KAPA HyperPrep, NEBNext Ultra II | Fragmentation, adapter ligation, and amplification of libraries | Reduced hands-on time, pre-mixed reagents, stable at room temperature |
| Automated Liquid Handling Consumables | Low-retention tips, pre-filled reagent plates, magnetic beads | Precise liquid transfers and purification steps | Compatibility with automated systems, reduced bubble formation, minimal retention |
| Quality Control Reagents | Qubit assay kits, Bioanalyzer reagents, qPCR quantification mixes | Assessment of nucleic acid quality, quantity, and library integrity | Pre-diluted standards, reduced pipetting steps, multi-plate compatibility |
| Normalization and Pooling Buffers | TE buffer, resuspension buffers, hybridization buffers | Standardization of library concentrations and preparation for sequencing | Chemical stability, viscosity optimization for automated pipetting |
| Sequencing Reagents | Illumina SBS chemistry, NovaSeq XP kits | Cluster generation and sequencing-by-synthesis | Enhanced stability, reduced volume requirements, increased output |
The growing demand for scalable genomic analysis represents a fundamental shift in biomedical research, particularly in high-throughput chemogenomics where the systematic profiling of compound-genome interactions drives therapeutic discovery. The market drivers analyzed in this application note – including technological advancements, expanding applications, and the adoption of cloud computing and AI – collectively underscore the critical importance of implementing automated, robust workflows for genomic analysis.
The protocols detailed herein provide a framework for laboratories seeking to enhance their throughput and reproducibility in chemogenomic studies. As the field continues to evolve, several emerging trends warrant particular attention. The integration of artificial intelligence into genomic analysis pipelines is accelerating, with models specifically designed for genomic data showing promise in predicting compound-gene interactions and optimizing experimental design [4]. Additionally, the continued expansion of multi-omics approaches will likely further drive demand for scalable solutions that can integrate genomic, transcriptomic, proteomic, and metabolomic data into a unified analytical framework [2].
For research organizations and drug development companies, strategic investment in automated NGS workflows represents not merely a tactical improvement but a fundamental capability that will determine competitive advantage in the evolving landscape of precision medicine and chemogenomic discovery.
High-Throughput Screening (HTS) is a foundational technology in modern drug discovery and chemogenomics research, enabling the rapid testing of thousands to millions of chemical compounds against biological targets. However, traditional manual approaches to screening create significant bottlenecks that limit throughput, introduce error, and constrain the scale of research. Manual processes are notoriously labor-intensive, requiring precise pipetting, repeated wash steps, and time-sensitive manipulations that are difficult to maintain across large-scale experiments [10]. These limitations become particularly problematic in the context of Next-Generation Sequencing (NGS) workflows, where the complexity of sample preparation can undermine the revolutionary throughput of the sequencing technology itself.
The integration of automation addresses these fundamental constraints by transforming HTS from a resource-intensive process to an efficient, reproducible, and scalable research platform. Automated systems streamline experimental workflows, minimize human error, and maximize throughput and efficiency through sophisticated liquid handling robots, integrated robotic arms, and advanced data analysis software [11]. This paradigm shift is especially critical for quantitative HTS (qHTS) paradigms, where compounds are tested at multiple concentrations to generate comprehensive concentration-response curves, requiring maximal efficiency and miniaturization [12]. By overcoming manual limitations, automation enables researchers to focus on experimental design and data interpretation rather than repetitive manual tasks, accelerating the entire drug discovery pipeline.
Key Bottlenecks in Manual HTS Workflows
Labor Intensity and Human Error
Manual HTS processes require extensive hands-on time for precise pipetting, repeated wash steps, and time-sensitive manipulations. In NGS workflows, for example, manual sample preparation necessitates numerous pipetting steps that create opportunities for human error [10]. These errors can be amplified during subsequent PCR stages, potentially ruining entire experiments and wasting significant time and resources. The consistency of manual pipetting varies between researchers, leading to batch effects where technical factors rather than biological variables influence results [10]. Such batch effects can mask true biological differences and lead to incorrect conclusions, particularly problematic in large-scale chemogenomics studies where reproducibility is essential.
Time Constraints and Throughput Limitations
The high-throughput potential of modern screening and sequencing technologies is fundamentally limited by manual sample preparation speeds. Manual processes create a significant bottleneck as researchers must spend vast amounts of time on preparatory work rather than innovative research [10]. This limitation restricts laboratory capabilities for large-scale experiments and reduces the time available for experimental design and data analysis. In conventional screening operations, the decoupling of screening and drug development has created unique challenges that demand efficient, unattended screening capabilities, particularly in academic settings where resources may be limited [12].
Contamination Risks and Consistency Issues
Cross-contamination presents a substantial risk during manual HTS and NGS sample preparation, potentially leading to inaccurate results and data misinterpretation [10]. The risk is particularly high when processing multiple samples simultaneously, as improper handling can compromise entire experimental batches. Additionally, maintaining consistency across manual processes is challenging, especially when scaling experiments for large studies or clinical applications. Researcher-to-researcher variations in technique introduce variability that can affect data quality and reproducibility [10].
Table 1: Primary Bottlenecks in Manual HTS and NGS Workflows
| Bottleneck Category | Specific Challenges | Impact on Research |
|---|---|---|
| Labor Intensity | Precise pipetting, repeated wash steps, time-sensitive manipulations | High error rates, increased hands-on time, reduced productivity |
| Time Constraints | Limited processing speed, extensive hands-on requirements | Restricted throughput, delayed experiments, reduced scalability |
| Contamination Risks | Cross-contamination between samples, environmental exposure | Inaccurate results, data misinterpretation, failed experiments |
| Consistency Issues | Researcher-to-researcher variation, batch effects | Reduced reproducibility, compromised data quality, invalid conclusions |
Automation Solutions for HTS Bottlenecks
Integrated Robotic Screening Systems
Modern automated screening systems incorporate multiple components into unified platforms capable of storing compound collections, performing assay steps, and measuring various outputs without human intervention. These systems typically include peripheral units such as assay and compound plate carousels, liquid dispensers, plate centrifuges, and plate readers, all serviced by high-precision robotic arms [12]. For example, the system implemented at the NIH's Chemical Genomics Center (NCGC) features random-access online compound library storage carousels with a capacity of over 2.2 million samples, extremely reliable plate handling, innovative lidding systems, multifunctional reagent dispensers, and anthropomorphic arms for plate transport and delidding [12]. Such integration enables fully automated unattended screening in the 1,536-well plate format, dramatically increasing efficiency while reducing reagent use and human error.
Liquid Handling and Process Automation
Liquid handling robots serve as the workhorses of automated HTS, accurately transferring samples and compounds into assay plates with precision and efficiency unmatched by manual pipetting [11]. These robotic systems employ advanced technology to manipulate small liquid volumes across hundreds or thousands of wells simultaneously, ensuring reproducibility across experiments and minimizing inter-sample variability. In modern HTS laboratories, these robots are seamlessly integrated with other automated systems including plate readers, imaging devices, and data analysis software, creating a cohesive workflow where each component communicates and coordinates with others [11]. This integration minimizes downtime between assay steps and maximizes throughput, enabling researchers to screen large compound libraries more rapidly and with greater consistency.
Automated Data Management and Analysis
Advanced software platforms form a critical component of automated HTS, tracking experimental parameters, documenting results, and analyzing the extensive datasets generated during screening campaigns [11]. These platforms automate essential data processing tasks including signal quantification, dose-response curve fitting, and hit identification, enabling researchers to derive meaningful insights more rapidly. The integration of artificial intelligence and machine learning (AI/ML) technologies further enhances data analysis capabilities, with algorithms trained on screening data to identify additional hits and prioritize compounds for further validation based on predicted activity, off-target effects, and drug-likeness [13]. Automated data FAIRification (Findability, Accessibility, Interoperability, and Reuse) protocols, such as those implemented in tools like ToxFAIRy, convert high-throughput data into machine-readable formats that support reuse and meta-analysis [14].
Diagram 1: HTS Automation Overcoming Manual Limitations. This workflow illustrates how automated solutions address specific bottlenecks in manual HTS processes.
Application Notes: Implementing Automated HTS in Chemogenomics
Quantitative HTS (qHTS) for Concentration-Response Profiling
The quantitative HTS (qHTS) paradigm represents a significant advancement made possible through automation, wherein each library compound is tested at multiple concentrations to construct concentration-response curves (CRCs) and generate comprehensive datasets for each assay [12]. This approach mitigates the high false-positive and false-negative rates associated with conventional single-concentration screening by testing compounds across a approximately four-log range of concentrations in an efficient, automated manner. At the NCGC, implementation of qHTS on an integrated robotic system has enabled the generation of over 6 million CRCs from more than 120 assays within three years [12]. The practical implementation of qHTS for cell-based and biochemical assays across libraries of >100,000 compounds requires maximal efficiency and miniaturization, as well as the ability to easily accommodate different assay formats and screening protocols – all capabilities provided by advanced automation systems.
High-Content Screening with Transcriptomic Readouts
Automation has enabled the evolution from simple HTS to high-content screening (HCS) that incorporates multiparameter analysis, including transcriptomic readouts. HCS platforms utilize automated imaging systems and advanced image analysis algorithms to gather quantitative data from complex cellular images, analyzing thousands of cells per well to provide detailed information on cellular morphology, protein localization, and signaling pathway activity [11]. Recent developments in high-throughput RNA-seq technology have further enhanced these capabilities by adding transcriptome-wide information to screening outputs. Methods like Discovery-seq provide a cost-effective way to obtain high-quality transcriptomics data during compound screens, offering comprehensive analysis of genes and pathways affected by chemical treatments [11]. This integrated approach provides a much deeper layer of information that researchers can gather from their HTS campaigns, enabling more sophisticated assessment of mechanisms of action, toxicity, and off-target effects much earlier in the drug development pipeline.
Toxicity Screening and Profiling
Automated HTS approaches have been successfully implemented for broad toxic mode-of-action-based hazard assessment through integrated testing protocols. These systems combine the analysis of multiple assays into comprehensive hazard values, such as the Tox5-score, which integrates dose-response parameters from different endpoints and conditions into a final toxicity score [14]. Automated platforms can simultaneously assess multiple toxicity endpoints including cell viability, DNA damage, oxidative stress, and apoptosis across several time points and cell models. The resulting data supports clustering and read-across based on endpoint, timepoint, and cell line specific toxicity scores, enabling bioactivity-based grouping of chemicals and nanomaterials [14]. This automated, multi-parametric approach to toxicity screening provides a more nuanced and informative alternative to traditional single-endpoint testing, facilitating better safety assessment of new chemical entities.
Table 2: Automated HTS Applications in Chemogenomics Research
| Application Area | Automated Approach | Key Benefits |
|---|---|---|
| Quantitative HTS (qHTS) | Testing each compound at multiple concentrations using automated dilution series | Generates comprehensive concentration-response data, reduces false positives/negatives |
| High-Content Screening | Automated imaging systems with advanced image analysis algorithms | Multiparameter cellular analysis, detailed morphological and functional data |
| Transcriptomic Profiling | High-throughput RNA-seq integrated with compound screening | Pathway-level understanding of compound effects, earlier mechanism of action data |
| Toxicity Screening | Automated multi-endpoint testing across time points and cell models | Comprehensive hazard assessment, supports bioactivity-based grouping |
Protocols for Automated HTS Implementation
Protocol: Quantitative HTS (qHTS) Implementation
Objective: To implement a quantitative high-throughput screening approach that tests each compound at multiple concentrations for robust concentration-response profiling [12].
Materials:
- Integrated robotic screening system with compound storage carousels
- 1,536-well assay plates
- Liquid handling robots with solenoid valve dispensers
- Plate readers compatible with various detection technologies
- Compound library formatted as concentration series
Procedure:
- System Configuration: Ensure the automated system includes random-access compound storage with capacity for concentration-series plates. The NCGC system configuration provides 1,458 positions dedicated to compound storage and 1,107 positions for assay plates [12].
- Assay Plate Preparation: Program liquid handlers to dispense reagents and cells into 1,536-well plates. Miniaturization to this format is essential for efficiency and reagent conservation.
- Compound Transfer: Utilize a 1,536-pin array for rapid compound transfer from source plates to assay plates across the concentration series.
- Incubation Management: Coordinate plate movement between multiple incubators capable of controlling temperature, humidity, and CO₂ to maintain optimal assay conditions.
- Detection and Reading: Automate plate transfer to appropriate detectors (e.g., ViewLux, EnVision) based on assay readout requirements (fluorescence, luminescence, absorbance, etc.).
- Data Processing: Implement automated data capture and concentration-response curve fitting using specialized software algorithms.
Validation: Include control compounds with known activity on each plate to monitor assay performance and system operation. The qHTS approach should generate between 700,000 and 2,000,000 data points per full-library screen [12].
Protocol: Automated High-Content Toxicity Screening
Objective: To perform automated multi-parameter toxicity assessment using five complementary endpoints for comprehensive hazard evaluation [14].
Materials:
- Automated plate replicators, fillers, and readers
- Cell culture systems (e.g., BEAS-2B cells)
- Assay reagents: CellTiter-Glo (viability), DAPI (cell number), gammaH2AX (DNA damage), 8OHG (oxidative stress), Caspase-Glo 3/7 (apoptosis)
- Test compounds and reference chemicals
- Automated imaging and analysis systems
Procedure:
- Cell Seeding: Automate cell dispensing into assay plates using liquid handlers. Maintain consistency across replicates.
- Compound Exposure: Program robotic systems to apply test materials at 12 concentration points across 4 biological replicates.
- Time Point Management: Schedule automated processing at multiple time points (e.g., 0, 6, 24, 72 hours) to capture kinetic responses.
- Endpoint Measurement: Coordinate sequential measurement of five toxicity endpoints:
- Luminescence measurement for cell viability via ATP content
- Fluorescence imaging for cell number using DAPI staining
- Fluorescence detection for caspase-3 activation (apoptosis)
- Oxidative stress assessment via 8OHG staining
- DNA double-strand break quantification through γH2AX staining
- Data Integration: Automate data collection and processing through the ToxFAIRy Python module or similar tools to calculate Tox5-scores [14].
- FAIRification: Convert experimental data and metadata into standardized, machine-readable formats using automated workflows.
Validation: Include reference chemicals and nanomaterial controls in each screening batch. The protocol should generate approximately 58,368 data points per screening campaign [14].
Diagram 2: Automated HTS Protocol Workflow. This diagram outlines the key steps in a generalized automated HTS protocol, highlighting quality control checkpoints.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagent Solutions for Automated HTS
| Reagent/Technology | Function | Application Notes |
|---|---|---|
| CellTiter-Glo Assay | Luminescent measurement of ATP content for viability assessment | Compatible with automation, provides reproducible viability data [14] |
| Caspase-Glo 3/7 Assay | Luminescent measurement of caspase activation for apoptosis detection | Suitable for automated screening platforms, kinetic measurements possible [14] |
| DAPI Staining | Fluorescent DNA staining for cell enumeration | Requires automated imaging systems, provides cell count data [14] |
| gammaH2AX Staining | Immunofluorescent detection of DNA double-strand breaks | Essential for genotoxicity assessment, compatible with automated HCS [14] |
| 8OHG Staining | Detection of nucleic acid oxidative damage | Marker of oxidative stress, requires automated imaging [14] |
| Unique Dual Index (UDI) Adapters | Barcode samples for multiplexed NGS | Critical for color-balanced sequencing, reduces index hopping [15] |
| Toehold Probes | Double-stranded molecular probes for variant detection | Enables color-mixing strategies for multiplex variant detection [16] |
| iTaq Universal Probes Supermix | PCR reaction mixture for probe-based detection | Compatible with automated liquid handling, consistent performance [16] |
Automation technologies have fundamentally transformed high-throughput screening by systematically addressing the critical bottlenecks associated with manual approaches. Through integrated robotic systems, precision liquid handling, and sophisticated data analysis tools, automation enables researchers to overcome limitations in throughput, reproducibility, and scalability that previously constrained chemogenomics research. The implementation of automated qHTS paradigms, high-content screening with transcriptomic readouts, and multi-parameter toxicity profiling demonstrates how these technologies enhance both the efficiency and quality of screening data. As automation continues to evolve with advancements in AI-driven data analysis and increasingly sophisticated robotic systems, its role in enabling robust, reproducible, and scalable high-throughput screening will only expand, further accelerating drug discovery and chemogenomics research.
The shift from manual procedures to automated systems in next-generation sequencing (NGS) is a pivotal transformation in modern genomics, particularly for high-throughput chemogenomics research. Automated liquid handlers and integrated workstations minimize hands-on time, reduce user-to-user variability, and enhance reproducibility, which is critical for generating robust, high-quality data in drug discovery pipelines [17]. These technologies enable researchers to standardize complex, multi-step NGS library preparation workflows, thereby accelerating the transition from genomic data to actionable therapeutic insights.
This application note details the key technologies, protocols, and practical considerations for implementing automated NGS solutions. It provides a framework for selecting the appropriate automation level—from modular liquid handlers to fully integrated workstations—to meet specific research throughput, budget, and application requirements.
The Scientist's Toolkit: Automated NGS Platforms and Reagents
Successful implementation of an automated NGS workflow requires a combination of specialized hardware and optimized reagent kits. The table below catalogues essential research reagent solutions and their functions within the automated workflow.
Table 1: Essential Research Reagent Solutions for Automated NGS Workflows
| Item | Function | Example Kits & Notes |
|---|---|---|
| Library Prep Kits | Fragments DNA/RNA and attaches sequencing adapters. | Illumina DNA Prep [18]; KAPA Library Prep kits [17]. Designed with overage for automated dead volumes. |
| Enrichment Panels | Selectively captures genomic regions of interest. | Used in Illumina DNA Prep with Enrichment [18]; crucial for targeted sequencing in chemogenomics. |
| Barcoding/Indexing Oligos | Uniquely tags individual samples for multiplexing. | Enables pooling of hundreds of samples [19]; critical for deconvolution in high-throughput screens. |
| Bead-Based Cleanup Reagents | Purifies nucleic acids between reaction steps. | G.PURE NGS Clean-Up Device [19]; automates removal of enzymes, primers, and adapter dimers. |
| Quantification Kits | Measures library concentration and quality. | Used pre-sequencing to ensure optimal loading [17]; can be integrated on-deck in some workstations. |
Commercially Available Automation Platforms
The market offers a spectrum of automation solutions, from flexible liquid handlers that can be incorporated into existing workflows to fully integrated, application-specific workstations. The choice depends on required throughput, level of walk-away automation, and budget.
Table 2: Comparison of Automated Liquid Handling Systems and Integrated Workstations
| Platform Type | Example Systems | Key Features | Throughput & Applications |
|---|---|---|---|
| Modular Liquid Handlers | Hamilton NGS STAR [18], Beckman Biomek i7 [18], Agilent Bravo NGS [20] | Flexible, open systems. Bravo NGS offers a compact design with optional on-deck thermal cycler (ODTC) [20]. | DNA Prep (96 libraries) [18]; RNA Prep (48 libraries) [18]. Ideal for labs with variable protocols. |
| Integrated Workstations | Aurora VERSA NGLP [21], Revvity explorer G3 [22], Roche AVENIO Edge [17] | Walk-away, end-to-end solutions. VERSA NGLP automates extraction, library prep, and PCR setup [21]. AVENIO Edge requires minimal setup time [17]. | Full workflow from nucleic acid extraction to ready-to-sequence libraries [21]. Best for standardized, high-volume labs. |
| Specialized & Low-Volume Systems | DISPENDIX I.DOT [19] | Non-contact dispenser for miniaturization. Capable of normalizing and pooling samples with a 1 µL dead volume [19]. | Dispenses nanoliter volumes; enables reaction miniaturization to 1/10th of standard volumes [19]. |
Detailed Experimental Protocols for Automated NGS
Protocol: Automated Library Preparation for Whole Genome Sequencing
Application Note: This protocol describes the automation of Illumina DNA Prep on a Hamilton Microlab NGS STAR or Beckman Biomek i7 liquid handler, enabling the preparation of 96 DNA libraries with over 65% less hands-on time compared to manual methods [18].
Materials:
- Input: 100 ng of high-quality genomic DNA per sample.
- Consumables: Illumina DNA Prep Kit [18], 96-well reaction plates, recommended tips.
- Equipment: Hamilton Microlab NGS STAR or Beckman Biomek i7 automated liquid handling system.
Methodology:
- Tagmentation: The automated system dispenses genomic DNA and tagmentation mix into a 96-well plate. The plate is transferred to an on-deck or external thermal cycler for a 10-minute incubation at 55°C to fragment the DNA.
- Stop Tagmentation and Amplification Prep: The system adds a neutralizing reagent to stop the tagmentation reaction, followed by the addition of a unique combination of Illumina CD Indexes to each sample well for sample multiplexing [18].
- PCR Amplification: The plate undergoes PCR cycling (program: 68°C for 1 minute; 98°C for 45 seconds, then cycle between 98°C for 15 seconds, 60°C for 30 seconds, and 68°C for 30 seconds for a total of 6-13 cycles; final extension at 68°C for 1 minute).
- Bead-Based Cleanup: The protocol integrates multiple bead-based cleanups using reagents like the G.PURE NGS Clean-Up Device to purify the DNA after tagmentation and PCR, removing enzymes, salts, and unwanted fragments [19].
- Final Library Elution: The purified DNA libraries are eluted in a resuspension buffer. The automated system can then proceed to normalization and pooling, or the libraries can be stored at -20°C.
Quality Control:
- Quantify final libraries using a fluorometric method (e.g., Qubit).
- Assess library size distribution using an instrument such as the Fragment Analyzer system [18].
Protocol: Automated Normalization and Pooling of NGS Libraries
Application Note: This protocol utilizes the DISPENDIX I.DOT Liquid Handler to normalize and pool up to 96 finished NGS libraries in a single, rapid step, minimizing dead volume and cross-contamination risk [19] [23].
Materials:
- Input: Quantified NGS libraries in a 96-well plate.
- Consumables: Fresh, clean source plate for the pooling destination.
- Equipment: DISPENDIX I.DOT Liquid Handler.
Methodology:
- Data Input: The concentration of each library, as determined by quality control, is input into the I.DOT software.
- Automated Normalization: The I.DOT calculates the volume required from each library to achieve an equimolar concentration. It then aspirates the libraries from the source plate.
- Simultaneous Pooling: The system dispenses the calculated, variable volumes of each normalized library into a single well of the destination plate. This combines the normalization and pooling steps into one efficient process [19].
- Verification: The I.DOT's software provides a log file confirming the volumes dispensed from each well, offering a record that the correct libraries were pooled as intended [19].
Technical Considerations for Implementation
Implementing an automated NGS workflow requires careful planning beyond selecting a platform. Key technical considerations ensure operational success and a strong return on investment.
- Throughput and Scalability: The system must align with your lab's current and projected sample volume. Integrated workstations like the Revvity explorer G3 are designed for high-throughput environments, whereas modular systems offer more flexibility for evolving needs [22].
- Liquid Handling Precision: For reaction miniaturization and costly clinical samples, precision at the nanoliter scale is paramount. Systems like the I.DOT with DropDetection technology verify dispensing volumes to ensure accuracy and result reliability [23].
- Software and Integration: The platform's software should be intuitive and compatible with Laboratory Information Management Systems (LIMS) for seamless sample tracking [23]. Evaluate the level of vendor support for protocol setup and troubleshooting, which is a key offering from providers like Illumina and Roche [18] [17].
- Cost and Return on Investment (ROI): While the initial investment is significant, automation provides long-term savings through reduced reagent consumption (via miniaturization), increased personnel efficiency, and higher data quality [23].
Workflow Diagram of an Automated NGS Pipeline
The following diagram illustrates the core stages of a fully automated NGS workflow, from sample input to sequencing-ready pools, and highlights the technologies involved at each step.
Diagram 1: Automated NGS workflow from sample to sequence-ready pool.
Automated liquid handlers and integrated workstations are no longer luxuries but core technologies for efficient, reproducible, and high-throughput NGS in chemogenomics research. The landscape of solutions is diverse, ranging from flexible platforms that automate specific protocol steps to walk-away systems that manage the entire workflow from sample extraction to sequencing-ready pools.
Selecting the right system requires a careful assessment of throughput needs, precision requirements, and the balance between initial investment and long-term efficiency gains. By leveraging validated protocols and partnering with vendors that offer robust application support, research teams can successfully deploy these key technologies to accelerate drug discovery and the development of novel therapeutics.
The escalating demand for reproducible, high-throughput data in chemogenomics and biomarker discovery is fundamentally reshaping next-generation sequencing (NGS) workflows. No single company possesses the complete suite of technologies required to seamlessly bridge the gap from biological sample to analyzable genomic data. This necessity has catalyzed a series of strategic partnerships between leading reagent providers and automation specialists. These collaborations are engineered to create integrated, end-to-end solutions that mitigate manual processing errors, enhance experimental reproducibility, and accelerate the pace of genomic discovery. This application note details specific partnerships and their resulting automated protocols, providing a framework for their implementation in high-throughput chemogenomics research.
The following table summarizes recent strategic partnerships that are defining the landscape of automated NGS workflows. These collaborations pair best-in-class assay chemistry with precision automation to address critical bottlenecks in sample preparation.
Table 1: Strategic Partnerships in Automated NGS Workflows
| Reagent Company | Automation Company | Collaborative Focus & Integrated Products | Key Benefits for Research | Status/Date |
|---|---|---|---|---|
| Integrated DNA Technologies (IDT) [24] [25] | Beckman Coulter Life Sciences [24] | Automation of IDT's Archer FUSIONPlex, VARIANTPlex, and xGen Hybrid Capture workflows on the Biomek i3 Benchtop Liquid Handler [24]. | Compact footprint; on-deck thermocycling; reduced hands-on time for lower-throughput sample volumes [24]. | In development (Nov 2025) [24]. |
| Integrated DNA Technologies (IDT) [25] | Hamilton [25] | Automation scripts for IDT's xGen and Archer NGS products on Hamilton's Microlab STAR and NIMBUS platforms [25]. | Scalability, consistency, and efficiency for comprehensive genomic profiling (CGP) in solid tumor and heme research [25]. | Global agreement (Oct 2025) [25]. |
| New England Biolabs (NEB) [26] | Volta Labs [26] | Integration of NEBNext reagents, starting with the Ultra II FS DNA Library Prep Kit, onto the Callisto Sample Preparation Platform [26]. | Fully automated, walk-away library prep; "Any Sequencer, Any Chemistry" flexibility (Illumina, Oxford Nanopore, PacBio) [26]. | Co-development partnership (Nov 2025) [26]. |
| HP [27] | Tecan [27] | Development of the Duo Digital Dispenser, combining single-cell and reagent dispensing using HP's inkjet technology [27]. | 40x faster drug discovery dosing; single-cell isolation in <5 minutes; surfactant-free reagent dispensing [27]. | Launched (May 2025) [27]. |
Detailed Experimental Protocols
Protocol: Automated Library Prep using IDT xGen Workflow on Hamilton NIMBUS Platform
This protocol outlines the procedure for automated library preparation for whole-genome sequencing using IDT's xGen hybridization capture reagents on a Hamilton NIMBUS system, derived from the stated partnership objectives [25].
The Scientist's Toolkit: Essential Materials
Table 2: Key Reagents and Consumables
| Item | Function / Description |
|---|---|
| IDT xGen Hybridization Capture Reagents [25] | A suite of probes designed for targeted sequencing, enabling the enrichment of specific genomic regions of interest. |
| Hamilton NIMBUS Liquid Handling Platform [25] | A precision automated workstation capable of performing complex liquid handling steps for NGS library construction. |
| NEBNext Ultra II FS DNA Library Prep Kit [26] | Provides enzymes and buffers for DNA fragmentation, end-prep, adapter ligation, and PCR amplification. |
| Microplates (96- or 384-well) | Reaction vessels compatible with the NIMBUS deck layout. |
| Magnetic Beads | For post-reaction clean-up and size selection steps. |
Methodology:
System Setup and Pre-Run Checklist:
- Ensure the Hamilton NIMBUS platform is calibrated. Load the validated method script developed through the IDT-Hamilton partnership [25].
- Position labware on the deck: source plates containing purified genomic DNA, reagent troughs with IDT xGen and NEB library prep reagents [25] [26], tip boxes, and microplate(s) for reactions.
- Pre-cool the on-deck thermocycler (if available).
Automated Fragmentation and End-Repair:
- The NIMBUS transfers a defined volume of genomic DNA (e.g., 100 ng in 50 µL) to the reaction plate.
- Using the integrated method, the system adds fragmentation mix from the NEBNext kit. The plate is transferred to the on-deck thermocycler for a programmed incubation to achieve desired fragment sizes (e.g., 300-400 bp) [26].
- Following fragmentation, the system adds end-repair enzyme mix to generate blunt-ended DNA fragments.
Adapter Ligation and Clean-Up:
- The workstation dispenses unique dual-indexed adapters and ligation master mix to each sample.
- After ligation, the protocol engages magnetic beads for a double-sided size selection clean-up. The NIMBUS performs all aspiration and dispensing steps to isolate the ligated product.
Hybridization Capture with IDT xGen Probes:
- The automated system transfers the purified library into a fresh plate and adds IDT xGen Hybridization Buffer and Blocking Oligos.
- The system then adds the specific xGen Probe Panels (e.g., for a cancer gene panel). The entire plate is sealed and transferred to the thermocycler for a prolonged hybridization incubation (e.g., 4-16 hours at 65°C) [25].
Post-Capture Amplification and Final Clean-Up:
- After hybridization, the system performs a series of stringent washes to remove non-specifically bound DNA.
- A post-capture PCR mix is added to amplify the enriched libraries.
- A final magnetic bead clean-up is performed. The NIMBUS elutes the final prepared library in elution buffer, ready for quantification and sequencing.
Workflow Visualization: Automated NGS from Sample to Sequencer
The following diagram illustrates the integrated, automated workflow described in the protocol, highlighting the roles of the respective partners' technologies.
Impact on Chemogenomics Research
The synergy from these partnerships delivers tangible benefits that directly address the core demands of high-throughput chemogenomics. Automated workflows ensure that the processing of hundreds of cell lines or compound-treated samples is consistent from batch to batch, a critical factor for generating robust, reproducible data for structure-activity relationship analysis [24] [25]. Furthermore, the significant reduction in hands-on time—a key benefit highlighted across all partnerships—frees highly skilled researchers to focus on experimental design and data interpretation rather than manual pipetting [24] [26]. Finally, the modular and scalable nature of these solutions, such as the "Any Sequencer, Any Chemistry" approach from Volta and NEB, provides the flexibility required to adapt to evolving research questions and sequencing technologies without overhauling core laboratory infrastructure [26].
Strategic collaborations between reagent and automation companies are more than a trend; they are a fundamental driver of innovation in modern genomics. By providing integrated, validated, and automation-ready workflows, these partnerships are directly empowering researchers to overcome traditional limitations of throughput, reproducibility, and scalability. As exemplified by the specific protocols and partnerships detailed herein, this collaborative model is proving indispensable for accelerating the pace of discovery in chemogenomics and the broader pursuit of precision medicine.
Building Your Automated NGS Workflow: From Library Prep to Data Generation
Automated liquid handling (ALH) systems and dedicated library preparation instruments are foundational to establishing robust, high-throughput Next-Generation Sequencing (NGS) workflows for chemogenomics research [28]. These technologies are critical for screening vast compound libraries against genomic targets, a process that demands exceptional precision, reproducibility, and scalability. The global NGS library preparation market, projected to grow from USD 2.07 billion in 2025 to USD 6.44 billion by 2034, reflects a significant shift towards automated and standardized workflows, with the automated preparation segment being the fastest-growing [29]. This application note provides a structured framework for selecting and implementing these core components to accelerate drug discovery.
Core Component Selection Criteria
Automated Liquid Handling Systems
ALH systems eliminate manual pipetting errors, reduce contamination risks, and standardize reagent dispensing, which is paramount for generating reliable, high-quality sequencing data in large-scale chemogenomics projects [28] [9]. When selecting a system, key features to consider include multi-channel pipetting, precision dispensing for sub-microliter volumes, and integration capabilities with Laboratory Information Management Systems (LIMS) [28].
Table 1: Key Considerations for Selecting an Automated Liquid Handler
| Consideration | Description & Relevance to Chemogenomics |
|---|---|
| Throughput Requirements | Dictates the scale of simultaneous processing. High-throughput systems are essential for screening large compound and genomic libraries [23]. |
| Precision and Accuracy | Critical for detecting single-nucleotide variants and ensuring data integrity in dose-response studies and genomic analysis [23]. |
| Sample Volume Ranges | The ability to accurately handle nanoliter volumes conserves precious clinical samples and high-value chemical compounds [23]. |
| Contamination Prevention | Features like disposable tips and acoustic liquid handling (non-contact) prevent cross-contamination between assay plates, ensuring result purity [23] [30]. |
| Integration with LIMS | Ensures full sample traceability from compound addition to sequencing data output, a key requirement for regulated research environments [28] [9]. |
Several types of liquid handling systems are available, each suited to different applications:
- Automated Liquid Handlers: Standard workhorses for most NGS library preparation steps, offering a balance of speed, accuracy, and flexibility [23].
- Integrated Workstations: Combine liquid handling with other instruments (e.g., plate sealers, washers) to create a fully automated walk-away solution for the entire library prep workflow [23].
- Acoustic Liquid Handlers: Use sound energy to transfer nanoliter-volume droplets without physical contact, ideal for miniaturized assays, PCR setup, and transferring precious samples in high-density plates [30].
Library Preparation Instruments
The selection of a library preparation platform directly impacts sequencing success. Automation in this stage standardizes processes, increases throughput, and enhances reproducibility by eliminating batch-to-batch variations inherent in manual protocols [9].
Table 2: NGS Library Preparation Market Overview & Trends (Data sourced from [29])
| Parameter | Market Data and Trends |
|---|---|
| Market Size (2025) | USD 2.07 Billion |
| Projected Market Size (2034) | USD 6.44 Billion |
| CAGR (2025-2034) | 13.47% |
| Dominating Region (2024) | North America (44% share) |
| Fastest Growing Region | Asia Pacific (CAGR of 15%) |
| Largest Product Segment | Library Preparation Kits (50% share in 2024) |
| Fastest-Growing Prep Type | Automated/High-Throughput Preparation (CAGR of 14%) |
Key technological shifts influencing instrument selection include the automation of workflows for higher efficiency and reproducibility, the integration of microfluidics for precise microscale control and reagent conservation, and advancements in single-cell and low-input kits that expand applications in oncology and personalized medicine [29].
Experimental Protocols for Automated NGS Workflows
Automated NGS Library Preparation Protocol
This protocol is designed for an integrated ALH system or workstation to process 96 samples for Illumina short-read sequencing.
Reagent Solutions:
- Fragmentation Mix: Enzymes or reagents for shearing DNA into desired fragment sizes.
- End-Repair & A-Tailing Mix: Enzymes to create blunt-ended, 5'-phosphorylated fragments with a single A-overhang for adapter ligation [31].
- Ligation Mix: Contains T4 DNA Ligase and indexing adapters with T-overhangs for ligation to A-tailed fragments [31].
- PCR Master Mix: Contains DNA polymerase, dNTPs, and primers for amplifying the final library.
- SPB Beads: Solid-phase reversible immobilization (SPRI) beads for post-reaction clean-up and size selection.
Procedure:
- DNA Normalization & Fragmentation:
- The ALH system transfers a calculated volume of each 50-200 ng DNA sample from a source plate to a 96-well PCR plate.
- The system then dispenses the Fragmentation Mix to each well.
- The plate is sealed, briefly centrifuged, and incubated on a thermal cycler as per the kit protocol to achieve the desired fragment size (e.g., 300-500 bp).
End-Repair & A-Tailing:
- The ALH system adds the End-Repair & A-Tailing Mix directly to the fragmented DNA.
- The plate is mixed, sealed, and incubated on a thermal cycler to create library-ready fragments.
Adapter Ligation:
- The ALH system dispenses a unique Ligation Mix containing a barcoded adapter into each well.
- The plate is incubated to allow adapters to ligate to the A-tailed fragments. This step enables sample multiplexing.
Post-Ligation Clean-Up:
- The ALH system adds a calibrated volume of SPB Beads to bind DNA.
- After incubation, the plate is placed on a magnetic stand. The ALH system, synchronized with the magnet, aspirates and discards the supernatant.
- Ethanol wash buffer is added and aspirated while the DNA-bound beads remain immobilized.
- Elution buffer is added, and the DNA is resuspended and eluted from the beads.
Library Amplification (PCR):
- The ALH system transfers the cleaned-up ligation product to a fresh PCR plate.
- A PCR Master Mix is dispensed into each well.
- The plate undergoes thermal cycling for a limited number of cycles to enrich for adapter-ligated fragments.
Final Library Clean-Up & Normalization:
- A final bead-based clean-up is performed using SPB Beads as in Step 4 to remove PCR reagents and primers.
- The final library is eluted in elution buffer or nuclease-free water.
- The ALH system can pool (multiplex) the libraries by transferring equal volumes based on prior quantification.
Quality Control:
- Quantify the final library using a fluorometric method (e.g., Qubit).
- Assess library size distribution using an instrument like the Agilent Bioanalyzer or TapeStation.
Diagram 1: Automated NGS Library Prep Workflow.
Protocol for Quantitative PCR Setup using Acoustic Liquid Handling
This protocol utilizes an acoustic liquid handler (e.g., Labcyte Echo) to miniaturize qPCR reactions for library quantification, significantly reducing reagent costs.
Reagent Solutions:
- qPCR Master Mix: Contains SYBR Green or TaqMan chemistry, DNA polymerase, dNTPs, and primers.
- Library Standards: A serially diluted library of known concentration to generate a standard curve.
- Nuclease-free Water: For diluting libraries and master mix.
Procedure:
- Plate Setup:
- Load a source plate containing the qPCR Master Mix onto the Echo.
- Load a separate plate with diluted Library Standards and experimental libraries.
- A low-volume destination qPCR plate (e.g., 384-well) is placed on the deck.
Reagent Transfer:
- The Echo uses sound energy to transfer precisely 2 µL of the qPCR Master Mix from the source plate to each well of the destination plate.
- Subsequently, it transfers 3 nL of each library (standard and unknown) from the sample plate into the corresponding wells containing the master mix. This non-contact transfer eliminates tip-based contamination and waste [30].
Sealing and Centrifugation:
- The destination plate is sealed with an optical film and briefly centrifuged to mix the contents and collect liquid at the bottom of the wells.
qPCR Run:
- The plate is transferred to a real-time PCR instrument and run according to the standard qPCR cycling protocol.
Data Analysis:
- The instrument software generates a standard curve from the known standards, which is used to calculate the concentration of each experimental library.
Implementation Strategy
Integration with Chemogenomics Workflow
For high-throughput chemogenomics, automated NGS components must function as part of a larger, integrated system. The library preparation process is a key step between target identification/compound treatment and bioinformatic analysis.
Diagram 2: NGS in High-Throughput Chemogenomics.
Quality Control and Compliance
Implementing real-time quality control is essential. Automated systems can be integrated with QC software (e.g., omnomicsQ) to monitor sample quality against pre-set thresholds, flagging failures before sequencing [9]. For drug development, adherence to regulatory standards like ISO 13485 and IVDR is critical. Automated systems support this compliance by ensuring standardized, documented, and reproducible workflows, facilitating participation in External Quality Assessment (EQA) programs [9].
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Automated NGS
| Item | Function in the Workflow |
|---|---|
| Library Preparation Kits | Pre-formulated reagent sets optimized for specific applications (e.g., exome, RNA-seq, single-cell). They ensure protocol consistency and high performance [29]. |
| Enzymes (Polymerases, Ligases) | Catalyze key reactions like PCR amplification and adapter ligation. Their quality and activity are critical for library yield and accuracy [31]. |
| Barcoded Adapters | Short DNA sequences ligated to fragments that enable sample multiplexing (pooling) on the sequencer and platform binding [31]. |
| Solid-Phase Reversible Immobilization (SPRI) Beads | Magnetic beads used for automated post-reaction clean-up, size selection, and buffer exchange throughout the library prep process. |
| Lyophilized Reagents | Stable, room-temperature reagents that remove cold-chain shipping and storage constraints, improving workflow flexibility and sustainability [29]. |
Next-generation sequencing (NGS) has revolutionized genomics, oncology, and infectious disease research, providing unprecedented insights into human health and disease [32]. However, manual NGS sample preparation presents significant challenges, including labor-intensive pipetting, sample variability, and reagent waste, creating critical bottlenecks in high-throughput chemogenomics research and modern laboratories [32]. The rising demand for NGS in clinical diagnostic settings, particularly for identifying genetic variations, diagnosing infectious diseases, and characterizing cancer mutations, necessitates solutions that ensure reproducible, reliable, and cost-effective results [32].
End-to-end automation addresses these challenges by transforming NGS workflows into streamlined, walk-away operations. Automated sample-to-sequencing platforms enhance data quality, improve operational efficiency, and enable scalable throughput while maintaining regulatory compliance [9] [33]. This application note details the implementation of fully automated NGS workflows within the context of high-throughput chemogenomics research, providing detailed protocols, performance metrics, and practical considerations for researchers and drug development professionals seeking to establish robust, hands-free sequencing operations.
The Case for Automation in NGS Workflows
Limitations of Manual NGS Processing
Manual NGS library preparation introduces multiple variables that compromise data quality and operational efficiency. Inconsistent pipetting techniques, sample tracking errors, and contamination risks during manual handling directly impact sequencing results [9]. These inconsistencies lead to poor quality outcomes that often require repetition, consuming additional time, financial resources, and precious samples [32]. Manual protocols also create substantial personnel burdens, with the Illumina DNA Prep protocol requiring approximately 3 hours of hands-on time per 8 samples processed [33].
The inherent variability of manual techniques poses particular challenges for chemogenomics research, where reproducible compound screening across large sample sets is essential for identifying therapeutic candidates. Batch-to-batch variation and differences in sample handling among staff members further complicate data interpretation and cross-study comparisons [34].
Advantages of Automated Platforms
Implementing end-to-end automation generates significant benefits across multiple dimensions of NGS operations:
Enhanced Data Quality and Reproducibility: Automated platforms perform precise liquid handling with minimal variability, producing consistent high-quality libraries [33]. Studies demonstrate that automation improves key NGS metrics including percentage of aligned reads, tumor mutational burden scoring, and median exon coverage [35]. This consistency is crucial for regulatory compliance in diagnostic applications and reliable compound screening in chemogenomics.
Substantial Time Savings: Automation dramatically reduces hands-on time while maintaining similar overall processing time. For example, automating the TruSight Oncology 500 assay reduced manual labor from approximately 23 hours to just 6 hours per run – a nearly four-fold decrease [35]. This efficiency gain allows researchers to focus on data analysis and experimental design rather than repetitive manual tasks.
Increased Throughput and Scalability: Automated systems can process 4 to 384 samples per run depending on the platform configuration, enabling laboratories to scale their sequencing operations without proportional increases in staffing [33]. This scalability is essential for chemogenomics applications that require screening large compound libraries across multiple cellular models.
Cost Optimization: While initial instrumentation investment is required (ranging from $45,000 to $300,000 depending on the system) [33], automation reduces long-term costs by minimizing reagent waste through precise nanoliter-range dispensing and decreasing failed runs due to human error [32] [34]. One study demonstrated that automated workflows can process thousands of samples weekly at less than $15 per sample [32].
Table 1: Quantitative Benefits of NGS Workflow Automation
| Performance Metric | Manual Process | Automated Process | Improvement |
|---|---|---|---|
| Hands-on time (TruSight Oncology 500) | ~23 hours/run | ~6 hours/run | 74% reduction [35] |
| Total process time | 42.5 hours | 24 hours | 44% reduction [35] |
| Aligned reads | ~85% | ~90% | ~5% increase [35] |
| Sample processing cost | Variable | <$15/sample | Significant cost reduction [32] |
| PCR hands-on time reduction | 3 hours | <15 minutes | >75% reduction [32] |
Integrated Automated NGS Platform Components
System Architecture for Walk-Away Operation
A fully automated NGS workstation integrates specialized instruments that perform complementary functions within the sequencing workflow. The G.STATION NGS Workstation exemplifies this integrated approach, incorporating the I.DOT Liquid Handler for non-contact reagent dispensing and the G.PURE NGS Clean-Up Device for magnetic bead-based purification [32]. This configuration enables complete walk-away operation for DNA-seq, RNA-seq, and targeted sequencing workflows.
Liquid handling systems form the core of automated NGS platforms, with major vendors including Hamilton, Beckman Coulter, Eppendorf, Tecan, and Revvity offering Illumina-compatible systems [18]. These systems provide precise fluid transfer across 96-, 384-, and 1536-well plate formats, enabling assay miniaturization that preserves precious reagents and samples [32]. The I.DOT Liquid Handler specifically dispenses in the nanoliter range, significantly reducing reagent consumption while maintaining assay integrity [32].
Integrated platforms incorporate ancillary modules that eliminate manual intervention points:
- On-deck thermocyclers for temperature-controlled incubations
- Magnetic bead handlers for purification and size selection steps
- Robotic arms for transferring plates between modules
- Barcode scanners for sample tracking and chain-of-custody documentation
This comprehensive integration enables true walk-away operation from sample preparation through sequencing-ready libraries.
Workflow Management and Quality Control
Automated NGS systems employ sophisticated software that orchestrates the entire sequencing workflow while monitoring quality parameters in real-time. Laboratory Information Management Systems (LIMS) integration enables complete sample tracking from nucleic acid extraction through library preparation, ensuring traceability for regulatory compliance [9].
Quality control software like omnomicsQ provides real-time monitoring of genomic samples, automatically flagging specimens that fail to meet pre-defined quality thresholds before they progress to sequencing [9]. This proactive quality assessment prevents wasted sequencing resources on suboptimal libraries and ensures only high-quality data advances through the pipeline.
Automated platforms also facilitate compliance with evolving regulatory frameworks including IVDR, ISO 13485, and ACMG guidelines [9]. The systems maintain detailed electronic records of all process parameters, reagent lots, and quality metrics necessary for diagnostic validation and audit trails.
Application Notes: Automated NGS in Chemogenomics Research
High-Throughput Compound Screening
Chemogenomics research requires systematic screening of chemical compounds against biological targets to identify therapeutic candidates. Automated NGS platforms enable comprehensive transcriptomic profiling of compound treatments at scales impractical with manual methods. Researchers can process hundreds of compound-treated samples in single runs, generating uniform RNA-seq libraries that reveal gene expression changes, alternative splicing events, and novel transcripts.
The I.DOT Liquid Handler has been specifically optimized for multiplex sequencing library preparation from low-input samples, enabling large-scale genomic surveillance applications [32]. This capability is particularly valuable for chemogenomics studies where sample material may be limited, such as primary cell cultures or patient-derived organoids. Automated systems can routinely process 48 DNA and 48 RNA samples simultaneously, compressing a 42.5-hour manual workflow into 24 hours [35].
Quality Control in Automated Screening
Maintaining quality standards across large compound screens presents significant challenges. Automated NGS platforms address this through integrated quality control checkpoints that assess RNA integrity, library concentration, and fragment size distribution at critical workflow stages. Systems can be programmed to automatically divert failing samples or flag them for review, preventing compromised libraries from consuming valuable sequencing resources.
Reference standards, including mock microbial communities or samples with well-defined sequence profiles, should be incorporated into each run to monitor workflow performance [34]. These controls enable continuous verification of sample lysis efficiency, nucleic acid extraction, cDNA synthesis, and overall library quality throughout automated operations.
Protocols for Automated NGS Library Preparation
Automated Whole Transcriptome Library Preparation
This protocol describes automated library preparation for RNA sequencing applications using the Hamilton NGS STARlet system with Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus, achieving over 65% reduction in hands-on time compared to manual methods [18].
Pre-Run Setup and Instrument Preparation
- Laboratory Preparation: Ensure all work surfaces are decontaminated using RNase decontamination solution. Thaw all reagents completely and mix by vortexing. Centrifuge briefly to collect contents at tube bottoms.
- Instrument Setup: Power on the Hamilton NGS STARlet system and associated on-deck thermocycler. Initialize the system software and select the appropriate protocol for RNA library preparation. Perform liquid class calibration if required by manufacturer specifications.
- Deck Layout Configuration: Load the deck according to the established map:
- Position 1: 96-well PCR plate for sample input
- Position 2: Tip boxes (300 µL and 50 µL)
- Position 3: Reagent reservoir with fragmentation mix
- Position 4: Reagent reservoir with ligation mix
- Position 5: Reagent reservoir with SPRI beads
- Position 6: Reagent reservoir with 80% ethanol
- Position 7: Empty reservoir for liquid waste
- Position 8: Magnetic module for bead separation
- Reagent Preparation: Dilute enzymes and buffers according to manufacturer specifications. Aliquot reagents to minimize freeze-thaw cycles. Ensure all reagents are at room temperature unless otherwise specified.
Automated Protocol Steps
RNA Quality Assessment (Pre-Automation)
- Quantify RNA samples using fluorometric methods (e.g., Qubit RNA HS Assay)
- Assess RNA integrity via capillary electrophoresis (e.g., Fragment Analyzer RNA Quality Number)
- Only proceed with samples having RQN/RIN > 8.0 for optimal results
Ribosomal RNA Depletion and Fragmentation
- System transfers 100-1000 ng total RNA to reaction plate
- Adds rRNA removal probes and hybridization buffer
- Incubates at 68°C for 5 minutes followed by 37°C for 10 minutes
- Adds RNase H and incubates at 37°C for 30 minutes
- Transfers fragmentation buffer and heats to 94°C for 8 minutes
cDNA Synthesis and End Repair
- Adds First Strand Synthesis Act D Mix and incubates at 25°C for 10 minutes
- Transfers First Strand Synthesis Buffer and enzyme, incubates at 42°C for 30 minutes
- Adds Second Strand Synthesis Mix and incubates at 16°C for 60 minutes
- Transfers End Repair Mix and incubates at 30°C for 30 minutes
Adapter Ligation and Cleanup
- Adds Ligation Mix with unique dual indexes to each sample
- Incubates at 23°C for 30 minutes
- Transfers SPRI beads for cleanup (0.9X ratio)
- Washes twice with 80% ethanol on magnetic module
- Elutes in 25 µL Resuspension Buffer
Library Amplification and Final Cleanup
- Adds PCR Master Mix and index primers
- Performs PCR: 98°C for 45s; 10-15 cycles of 98°C for 15s, 60°C for 30s, 72°C for 30s; final extension at 72°C for 1 minute
- Performs final SPRI bead cleanup (0.9X ratio) with elution in 25 µL Resuspension Buffer
Post-Processing Quality Control
- Quantify libraries using fluorometric methods (e.g., Qubit dsDNA HS Assay)
- Assess library size distribution via capillary electrophoresis (e.g., Fragment Analyzer)
- Verify expected peak size of approximately 300-500 bp
- Pool libraries at equimolar ratios for sequencing
- Store final libraries at -20°C until sequencing
Automated Targeted Sequencing for Mutation Screening
This protocol describes automated preparation of targeted sequencing libraries using the Illumina DNA Prep with Enrichment on Beckman Biomek i7 systems, ideal for mutation screening in chemogenomics applications.
Specialized Reagent Setup
- Hybridization Capture Reagents: Prepare blocking oligonucleotides, biotinylated capture probes, and streptavidin-coated magnetic beads according to panel specifications.
- Indexing Strategy: Plan dual index combinations to enable sample multiplexing while preventing index hopping artifacts. Use unique combinations for each sample in large screens.
- Bead Preparation: Resuspend magnetic beads thoroughly by vortexing for at least 30 seconds or until fully homogenized.
Automated Targeted Capture Protocol
Library Preparation
- Follow steps for fragmentation, end repair, and adapter ligation as in section 5.1.2
- Use unique dual indexes for each sample during adapter ligation
Hybridization Capture
- Transfer 250-500 ng purified library to new plate
- Add hybridization buffer and pooled capture probes
- Incubate at 95°C for 10 minutes followed by 65°C for 16-24 hours
Target Selection
- Add streptavidin magnetic beads and incubate at 65°C for 45 minutes
- Wash beads twice with Wash Buffer I at room temperature
- Wash beads twice with Stringent Wash Buffer at 65°C
- Elute captured DNA in Elution Buffer at 95°C for 10 minutes
Capture Amplification
- Transfer eluted DNA to new plate containing PCR Master Mix
- Perform 10-12 amplification cycles
- Clean up with SPRI beads (0.9X ratio)
- Elute in 25 µL Resuspension Buffer
Quality Assessment for Targeted Libraries
- Verify library concentration (recommended > 10 nM)
- Confirm fragment size distribution (expected peak ~300-400 bp)
- Assess capture efficiency via qPCR if required
- Validate panel performance using positive control samples with known variants
Table 2: Automated NGS Protocol Performance Metrics
| Protocol | System | Hands-on Time | Total Processing Time | Sample Throughput | Key Applications |
|---|---|---|---|---|---|
| Whole Transcriptome | Hamilton NGS STARlet | <30 minutes | ~7 hours | 48 samples | Compound transcriptomics, differential expression [18] |
| Targeted Sequencing | Beckman Biomek i7 | ~1 hour | ~24 hours | 48-96 samples | Mutation screening, variant validation [18] |
| Whole Genome Sequencing | Hamilton/Beckman | <30 minutes | ~6 hours | 96 samples | Genomic variant discovery, structural variants [18] |
| Single-Cell RNA-seq | Biomek i7 | 45 minutes | ~8 hours | 96 samples | Cellular heterogeneity, compound effects [35] |
Research Reagent Solutions for Automated NGS
Successful implementation of automated NGS workflows requires specialized reagents formulated for robotic liquid handling systems. The following table details essential solutions optimized for automated sample-to-sequencing platforms.
Table 3: Essential Research Reagent Solutions for Automated NGS Workflows
| Reagent Category | Specific Products | Function in Workflow | Automation-Specific Features |
|---|---|---|---|
| Library Preparation Kits | Illumina DNA Prep, Illumina Stranded Total RNA Prep | Fragmentation, adapter ligation, library amplification | Reduced viscosity, optimized for precise non-contact dispensing [18] |
| Target Enrichment | Illumina DNA Prep with Enrichment, Twist Target Enrichment | Hybridization capture, amplicon generation | Compatible with automated bead-based cleanups, stable at room temperature [18] [35] |
| Magnetic Beads | SPRIselect, G.PURE NGS Clean-Up beads | Size selection, purification | Uniform size distribution, consistent binding capacity, rapid magnetic separation [32] |
| Enzyme Mixes | Watchmaker Genomics enzymes | Fragmentation, amplification, modification | Highly concentrated formulations, reduced glycerol content, stable at 4°C [35] |
| Liquid Handling Reagents | LowTE buffer, customized resuspension buffers | Dilution, normalization, storage | Optimized surface tension for accurate dispensing, non-foaming compositions [32] |
| Quality Control Kits | D1000 ScreenTape, Qubit dsDNA HS Assay | Quantification, size distribution analysis | Compatible with automated liquid handlers, minimal hands-on steps [18] |
Implementation Strategy and Operational Considerations
Platform Selection and Validation
Choosing the appropriate automation platform requires careful assessment of current and projected workflow needs. Key considerations include:
- Throughput Requirements: Match system capacity (samples per run) to screening scale with 20-30% overhead for future growth
- Application Diversity: Ensure platform supports diverse NGS methods (DNA-seq, RNA-seq, targeted panels) used in chemogenomics research
- Integration Capabilities: Verify compatibility with existing laboratory instruments and information systems
- Validation Requirements: Plan for comprehensive performance qualification using reference standards and comparison to established manual methods
Implementation should follow a phased approach, beginning with validation of individual workflow steps before progressing to complete end-to-end operation. Parallel testing of automated and manual methods using standardized reference samples establishes performance baselines and identifies potential optimization requirements.
Operational Excellence and Troubleshooting
Maintaining consistent performance of automated NGS platforms requires dedicated operational protocols:
- Preventive Maintenance: Establish regular calibration schedules for liquid handling components, with daily, weekly, and monthly maintenance tasks
- Quality Monitoring: Implement statistical process control for key performance indicators including library yield, success rates, and sequencing metrics
- Staff Training: Designate and train "super users" proficient in troubleshooting common errors, deck teaching, and protocol modifications [33]
- Contingency Planning: Maintain competency in manual preparation methods to ensure workflow continuity during instrument service intervals [33]
Common operational challenges include liquid handling errors due to reagent viscosity, magnetic bead separation inconsistencies, and sample plate positioning inaccuracies. Systematic troubleshooting protocols should document solutions for these recurrent issues to minimize system downtime.
End-to-end automation of NGS workflows represents a transformative advancement for high-throughput chemogenomics research. Integrated sample-to-sequencing platforms deliver reproducible, high-quality data while dramatically reducing hands-on time and operational costs. The implementation framework detailed in this application note provides a roadmap for establishing robust walk-away operations that accelerate drug discovery and development.
As NGS technologies continue evolving, strategic partnerships between reagent manufacturers and automation vendors will further streamline workflows and enhance capabilities [35]. Future developments in artificial intelligence-driven quality control, integrated multi-omics workflows, and predictive analytics will expand the role of automated NGS platforms in chemogenomics research, enabling more sophisticated compound screening and mechanistic studies.
Successful implementation requires careful platform selection, systematic validation, and ongoing performance monitoring, but delivers substantial returns through increased throughput, enhanced data quality, and operational efficiency. By adopting these automated systems, research organizations can position themselves at the forefront of genomic science and therapeutic discovery.
Assay miniaturization is the process of scaling down reaction volumes while maintaining the accuracy and precision of standard-volume assays [36]. This approach has become fundamental across drug discovery, diagnostics, and personalized medicine, enabling high-throughput screening (HTS) in reduced volumes and facilitating more extensive compound testing with limited available compound volumes [36]. A critical enabling technology for successful miniaturization is non-contact dispensing, which allows for precise liquid handling without direct contact with reagents or substrates, thereby minimizing contamination risks and conserving valuable reagents [37] [38]. Within the context of automated next-generation sequencing (NGS) workflows for high-throughput chemogenomics research, miniaturization transforms laboratory practices by significantly reducing reagent costs, decreasing plastic waste, and increasing experimental throughput [39] [40]. The integration of non-contact dispensers, such as the I.DOT Liquid Handler, provides the technological foundation for reliably miniaturizing complex biochemical reactions, including NGS library preparations, to volumes as low as one-tenth of manufacturer-recommended protocols without compromising data quality [41] [40].
Key Principles of Non-Contact Dispensing
Fundamental Technology and Mechanisms
Non-contact dispensing systems operate on principles fundamentally different from traditional liquid handlers. Instead of using air displacement mechanisms with disposable tips, these systems typically employ positive pressure pulses to eject droplets through precisely molded pores in disposable source wells [37]. Each droplet is formed and ejected without the dispensing mechanism touching the target well or the liquid itself. Advanced systems incorporate DropDetection sensors that verify the actual number of droplets dispensed, providing real-time process control and ensuring volumetric accuracy [37] [42]. This technology enables precise dispensing of volumes as low as 8 nL with resolution of 0.1 nL, making it particularly suitable for miniaturized assays where minute volume differences can significantly impact results [41].
Advantages Over Traditional Liquid Handling
The transition to non-contact dispensing offers several substantial advantages for automated laboratory workflows. By eliminating the need for pipette tips, these systems dramatically reduce consumable costs and plastic waste, contributing to more sustainable laboratory operations [37] [40]. The non-contact nature of the technology virtually eliminates cross-contamination between samples, a critical consideration for sensitive NGS applications [37] [38]. Additionally, these systems exhibit extremely low dead volumes (as low as 1 μL per dispense), crucial for conserving expensive reagents in miniaturized protocols [41]. The operational efficiency is also significantly enhanced, with systems capable of dispensing 10 nL across a 384-well plate in approximately 20 seconds, enabling rapid high-throughput processing essential for chemogenomics research [37].
Quantitative Benefits of Miniaturization in NGS Workflows
Assay miniaturization, when implemented with non-contact dispensing technology, delivers measurable improvements in cost efficiency, sustainability, and operational performance. The following tables summarize key quantitative benefits observed in research settings.
Table 1: Cost and Time Savings from Workflow Miniaturization
| Parameter | Standard Protocol | Miniaturized Protocol | Reduction/Savings |
|---|---|---|---|
| Reagent Volume | Manufacturer's recommended volume (e.g., 20 μL) | 1/10th volume (e.g., 2 μL) | Up to 90% reduction [40] |
| Reagent Costs | Full price | Miniaturized consumption | Up to 86% savings [40] |
| Hands-on Time | Manual processing hours | Automated miniaturized workflow | Over 150 hours saved in NGS library prep [40] |
| Plastic Consumables | Standard tip usage | Tip-reduced or tipless workflow | Significant reduction in plastic waste [37] [40] |
Table 2: Performance Metrics of Non-Contact Dispensing Systems
| Performance Characteristic | Capability/Range | Significance |
|---|---|---|
| Volume Range | 8 nL to 30 μL [41] | Enables dramatic assay miniaturization |
| Dispensing Precision | CV of 0.5% to 5.3% [42] | Ensves reproducible results in miniaturized formats |
| Viscosity Compatibility | Methanol to 65% glycerol [41] | Handles diverse reagents without recalibration |
| Throughput | 384-source liquids per run [41] | Supports high-throughput screening requirements |
| Dead Volume | As low as 1 μL per dispense [41] | Maximizes reagent conservation |
Application Note: Miniaturized NGS Library Preparation for Chemogenomics
Background and Objective
Chemogenomics research requires the systematic screening of chemical compounds against biological targets to identify novel therapeutic candidates. NGS library preparation is a critical step in profiling cellular responses to compound treatments, but traditional protocols consume substantial quantities of expensive reagents and limit screening throughput. This application note details a miniaturized NGS library preparation protocol leveraging non-contact dispensing technology to achieve 90% reagent reduction while maintaining library quality and sequence data integrity for chemogenomics applications.
Experimental Protocol
Materials and Equipment
Table 3: Research Reagent Solutions for Miniaturized NGS Library Prep
| Item | Function | Considerations for Miniaturization |
|---|---|---|
| Fragmentation Mix | Fragments DNA/RNA to appropriate size | Volume reduction requires precise dispensing to maintain enzyme-to-substrate ratios [39] |
| End Repair & A-Tailing Enzymes | Prepares fragments for adapter ligation | Maintain activity at reduced volumes; cold handling may be required [41] |
| Ligation Mix with Barcoded Adapters | Adds platform-specific adapters | Critical for multiplexing; minimal dead volume essential for cost savings [39] |
| SPRI Beads | Size selection and purification | Magnetic bead handling optimized for small volumes [39] |
| PCR Master Mix | Amplifies final library | Enzyme stability must be maintained through potential temperature fluctuations [39] |
| Nuclease-free Water | Volume adjustment | Ultra-pure quality essential for reproducible results at low volumes [39] |
Methodology
Step 1: Fragmentation and Size Selection Begin with 10-100 ng of input DNA or RNA in a 2 μL volume. Add 0.5 μL of fragmentation mix using the non-contact dispenser with integrated volume verification. Incubate according to manufacturer recommendations but with reduced duration (typically 75% of standard time). Clean up using 0.8x SPRI beads in a 4 μL reaction volume, separating on a magnetic rack adapted for 384-well plates [39] [40].
Step 2: End Repair and A-Tailing Resuspend fragmented DNA in 3.5 μL of end repair and A-tailing master mix, dispensed using the non-contact dispenser. The precise formulation maintains enzyme concentration while reducing total volume 10-fold compared to standard protocols. Incubate at 20°C for 15 minutes followed by 65°C for 15 minutes [39].
Step 3: Adapter Ligation Add 1.5 μL of ligation mix containing molecularly barcoded adapters using the non-contact dispenser. Use reduced adapter concentrations (typically 0.5-1 μM final concentration) optimized for miniaturized reactions. Incubate at 20°C for 15 minutes [39] [40].
Step 4: Post-Ligation Cleanup and PCR Amplification Perform SPRI bead cleanup with 6 μL of beads in a total volume of 10 μL. Elute in 5 μL of nuclease-free water. For PCR amplification, prepare a 5 μL reaction containing 0.5-1 μL of eluted library, reduced primer concentrations, and a hot-start PCR master mix. Amplify with cycle number determined by input amount and library complexity requirements [39] [40].
Step 5: Library Quantification and Quality Control Quantify libraries using fluorescence-based methods compatible with low-volume measurements (e.g., 1 μL samples). Assess size distribution using microfluidic electrophoresis systems requiring only 1 μL of sample [39].
Workflow Visualization
Diagram 1: Miniaturized NGS library preparation workflow. Key miniaturization points (yellow nodes) represent steps where volume reduction is most significant.
Results and Discussion
Implementation of this miniaturized NGS library preparation protocol with non-contact dispensing technology demonstrated equivalent library quality to standard protocols while reducing reagent consumption by approximately 90% [40]. Sequencing metrics including library complexity, duplicate rates, and coverage uniformity showed no significant differences between miniaturized and standard protocols. The reduced reaction volumes enabled processing of four times more samples with the same reagent budget, dramatically increasing screening throughput for chemogenomics applications. The non-contact dispensing system maintained coefficients of variation below 5% for all liquid handling steps, ensuring reproducible results across 384-well and 1536-well formats [42] [41].
Implementation Considerations for Automated NGS Workflows
System Selection and Integration
When implementing non-contact dispensing for assay miniaturization, careful consideration of system capabilities is essential. The system should demonstrate proven compatibility with the viscosity range of reagents used in NGS workflows, from aqueous solutions to glycerol-containing enzymes [41]. Integration capabilities with existing laboratory automation should be assessed, including compatibility with SBS-standard labware, API accessibility for workflow automation, and physical footprint constraints [37] [41]. For temperature-sensitive reagents, optional cooling/heating modules maintain enzyme activity and reagent integrity during dispensing operations [41].
Protocol Optimization Strategies
Successful miniaturization requires more than simple volume reduction. Reagent concentrations may require optimization to maintain effective enzyme-to-substrate ratios in reduced volumes [39]. Incubation times can often be shortened due to reduced diffusion distances in smaller volumes [39]. Magnetic bead-based cleanups should be adapted for small volumes, potentially requiring adjustments to bead-to-sample ratios [39]. Each protocol should undergo rigorous validation against standard methods to ensure equivalent performance before implementation in high-value screening campaigns.
Economic Justification and Sustainability Impact
The economic justification for implementing non-contact dispensing extends beyond reagent savings. A comprehensive return-on-investment analysis should account for reduced consumable costs (pipette tips), increased throughput, and personnel time reallocation from manual liquid handling to higher-value activities [37] [40]. The environmental impact is substantial, with laboratories potentially reducing plastic waste by thousands of kilograms annually while simultaneously decreasing energy consumption through smaller instrument footprints [40].
Non-contact dispensing technology enables robust assay miniaturization that delivers substantial cost savings, enhanced sustainability, and increased throughput for automated NGS workflows in chemogenomics research. The precise volumetric control, minimal dead volume, and contamination-free operation of these systems make them indispensable tools for laboratories seeking to maximize research output while conserving valuable resources. As the field advances toward increasingly automated and integrated laboratory environments, non-contact dispensing will play a pivotal role in enabling the next generation of high-throughput genomic screening essential for drug discovery and development.
Next-generation sequencing (NGS) has revolutionized genomics by enabling rapid, high-throughput DNA and RNA analysis, with its ability to decode genetic information quickly and accurately transforming fields like medicine, agriculture, and environmental science [43]. As we approach 2025, automated NGS services are becoming more accessible and integrated into everyday workflows, driving innovation and efficiency across industries. The integration of automation technologies addresses critical bottlenecks in library preparation and data analysis, significantly reducing hands-on time from hours to minutes while improving reproducibility and consistency [44] [45]. This application note explores the implementation of automated NGS workflows across three critical domains—oncology, infectious disease surveillance, and single-cell analysis—within the broader context of high-throughput chemogenomics research.
For chemogenomics research, which involves systematic study of interactions between chemical compounds and biological systems, automated NGS workflows enable unprecedented scaling of experimental throughput. The ability to process hundreds of samples simultaneously with minimal human intervention accelerates target identification, mechanism of action studies, and compound efficacy assessment. This technical advancement is particularly valuable for drug development professionals seeking to establish robust, reproducible pipelines for preclinical research.
Automated NGS in Oncology
In oncology, automated NGS has become indispensable for precision medicine approaches, enabling detailed tumor profiling that helps oncologists tailor treatments to individual genetic profiles [43]. The identification of specific mutations through automated NGS panels allows clinicians to select targeted therapies, significantly improving patient outcomes. Adoption rates have shown a 30% increase in personalized treatment plans over the past three years, with automated workflows now routinely used to detect actionable mutations in lung and breast cancers [43]. The transition to automated systems addresses key challenges in oncology testing, including the need for consistent results, reduced workflow errors, and improved reproducibility across multiple laboratory settings [45].
Recent industry developments highlight the strategic importance of automation in oncology NGS. Clear Labs, for instance, has expanded its automation platform beyond infectious disease sequencing to oncology, announcing a collaboration with Labcorp to develop streamlined, oncology-focused NGS workflows [46]. This collaboration aims to develop end-to-end workflows that help laboratories improve efficiency, consistency, and throughput while laying the groundwork for future adoption across a wide range of genomic applications, with early access to automated oncology workflows planned for 2026 [46].
Experimental Protocol: Automated Solid Tumor Profiling
Objective: To identify actionable genomic alterations in solid tumor samples using an automated NGS workflow for therapeutic targeting.
Materials and Equipment:
- Clear Dx Automated NGS Platform or equivalent automated system
- FFPE tumor tissue sections (5-10 μm thick) with >20% tumor content
- Automated nucleic acid extraction system (e.g., chemagic 360)
- PGDx elio complete capture kit or equivalent oncology panel
- Liquid handling system with temperature control (e.g., Fontus workstation)
- Illumina NovaSeq X Plus sequencing platform or equivalent
- Bioinformatics pipeline for variant calling (e.g., CSI NGS Portal)
Methodology:
- Nucleic Acid Extraction: Process FFPE sections using automated extraction system according to manufacturer's protocol. Quantify DNA yield using fluorometric methods (e.g., Qubit Fluorometer).
- Library Preparation: Utilize automated liquid handling systems to fragment 50-100ng of input DNA, followed by end-repair, A-tailing, and adapter ligation. Employ unique molecular identifiers (UMIs) to correct for PCR and sequencing errors.
- Target Enrichment: Perform hybrid capture using oncology-specific panels covering 100-500 cancer-associated genes. Automate the hybridization and washing steps using temperature-controlled liquid handlers.
- Library Quantification and Normalization: Quantify libraries using qPCR-based methods (considered gold standard for NGS quantification) to ensure accuracy and sensitivity. Normalize libraries to 2nM concentration using automated systems like the Myra liquid handling system, which provides precision normalization critical for balanced representation.
- Pooling and Sequencing: Pool up to 24 normalized libraries in equimolar ratios and sequence on a high-throughput platform (e.g., Illumina NovaSeq X Plus) using 2×150 bp paired-end reads.
- Data Analysis: Process raw sequencing data through automated bioinformatics pipelines for:
- Alignment to reference genome (GRCh38) using BWA-MEM
- Mutation calling (SNVs, indels, CNVs, fusions) using optimized algorithms
- Annotation of variants using clinical databases (OncoKB, CIViC)
- Generation of clinical reports highlighting actionable alterations
Quality Control Measures:
- Implement PhiX control (1%) for sequencing quality monitoring
- Maintain cluster density between 200-300K/mm² with >80% passing filter
- Achieve Q-score >30 (equivalent to <0.1% error rate) across all bases
- Ensure >100x coverage depth for >95% of target regions
- Include positive control samples with known mutations in each run
Key Performance Metrics in Automated Oncology NGS
Table 1: Quantitative performance metrics for automated oncology NGS workflows
| Parameter | Performance Metric | Clinical Validation Threshold |
|---|---|---|
| Hands-on Time | Reduced from 8h to 45min for library prep [44] | >75% reduction compared to manual methods |
| Sample Throughput | Up to 384 samples processed simultaneously [44] | Minimum 96 samples per run |
| Turnaround Time | <48 hours from sample to report | <72 hours for clinical reporting |
| Reproducibility | CV <5% across samples and runs [47] | CV <10% for clinical applications |
| Sensitivity | >99% for variants at >5% allele frequency | >95% for variants at >5% allele frequency |
| Specificity | >99.9% for all variant types | >99.5% for all variant types |
| Cost per Sample | ~25% reduction compared to manual processing | Minimum 20% cost reduction |
Automated NGS in Infectious Disease Surveillance
The COVID-19 pandemic highlighted NGS's critical role in pathogen detection and tracking, establishing automated infectious disease surveillance as a vital public health tool [43]. In 2025, automated NGS continues to be essential for monitoring emerging infectious diseases, tracking mutations, and developing vaccines. Public health agencies utilize automated NGS for rapid outbreak investigation and pathogen characterization, with its ability to analyze complex microbial communities also aiding in antibiotic resistance studies [43]. Fully automated, end-to-end solutions like the Clear Dx System enable next-day identification of bacterial and fungal pathogens from sterile site specimens, significantly accelerating diagnostic timelines [45].
Environmental monitoring through wastewater sequencing has emerged as a powerful application of automated NGS for infectious disease surveillance. Systems like the Clear Dx FlexPro: Wastewater provide fully automated end-to-end whole genome sequencing solutions for monitoring pathogens like SARS-CoV-2 in wastewater, enabling early detection of community outbreaks [45]. Similarly, automated microbial surveillance WGS solutions support public health efforts in tracking bacterial and fungal isolates across healthcare settings [45].
Experimental Protocol: Automated Wastewater Pathogen Surveillance
Objective: To detect and characterize pathogen prevalence and variants in wastewater samples using automated NGS.
Materials and Equipment:
- Automated wastewater concentration system (centrifugation or filtration-based)
- Automated nucleic acid extraction system with pathogen inactivation capability
- Clear Dx Microbial ID tNGS assay workflow or equivalent automated platform
- Targeted amplification reagents for specific pathogens (e.g., SARS-CoV-2, Influenza)
- Liquid handling robots with 96- or 384-well capability
- Portable or benchtop sequencer (e.g., Oxford Nanopore MinION, Illumina iSeq 100)
- Cloud-based data analysis platform with automated reporting
Methodology:
- Sample Collection and Concentration: Automate the collection of 24-hour composite wastewater samples. Process 50-200mL of wastewater through centrifugation or membrane filtration systems to concentrate particulates.
- Nucleic Acid Extraction and Purification: Utilize automated extraction systems to simultaneously process 24-96 samples with integrated pathogen inactivation steps. Incorporate extraction controls to monitor efficiency and potential inhibition.
- Library Preparation: Employ automated NGS systems that integrate the following steps:
- cDNA synthesis for RNA viruses using random hexamers and target-specific primers
- Whole genome amplification using multiplex PCR approaches
- DNA fragmentation (if required for DNA pathogens)
- End-repair, A-tailing, and adapter ligation using automated liquid handling
- Library quantification via fluorometric methods integrated into the workflow
- Normalization and Pooling: Use automated systems like Myra to normalize libraries to 4nM concentration and pool up to 96 samples based on calculated volumes, ensuring equal representation. The system's level sensing feature detects air pockets in wells, minimizing sample drop-outs [47].
- Sequencing: Load pooled libraries onto appropriate sequencing platforms. For rapid turnaround, utilize portable systems; for higher throughput, use benchtop sequencers.
- Data Analysis and Reporting: Implement automated bioinformatics pipelines for:
- Quality control of raw reads (FastQC)
- Reference-based mapping (BWA, Bowtie2)
- Variant calling (GATK, iVar)
- Phylogenetic analysis and lineage assignment
- Automated report generation for public health decision-making
Quality Control Measures:
- Include process controls (e.g., mengovirus) to monitor extraction efficiency
- Implement amplification controls to detect PCR inhibition
- Sequence known reference samples for variant calling validation
- Maintain minimum coverage of 100x for variant identification
- Monitor read quality scores (Q>30) across all samples
Signaling Pathways in Host-Pathogen Interactions
Diagram 1: Host-pathogen interaction signaling pathways. Automated NGS can identify pathogen factors that modulate these pathways.
Automated NGS in Single-Cell and Spatial Analysis
Single-cell technologies, known collectively as single-cell and spatial omics, have opened a new frontier for cell biology by providing higher-resolution insight into cellular heterogeneity [44]. These technologies enable the analysis of the genome and transcriptome of large numbers of individual cells within a sample, allowing scientists to understand the nuanced impact of spatiotemporal organization of individual cells on everything from embryonic development to tumor progression and aging [44]. Single-cell transcriptomics enables researchers to investigate the expression profile of individual cells, uncovering the incredible heterogeneity that exists within tissues, while spatial transcriptomics provides an additional layer of information, revealing the spatial context of gene expression data within a tissue or organ [44].
The potential of these techniques to revolutionize drug discovery, personalized medicine, and our ability to unravel the molecular underpinnings of various diseases is profound, promising a future where targeted therapies and interventions are more precise and effective than ever before [44]. However, these advanced techniques require sophisticated automation solutions to overcome the significant challenges associated with manual library preparation workflows, which are laborious, time-consuming, and highly sensitive to variation and contamination [44].
Experimental Protocol: Automated Single-Cell RNA Sequencing
Objective: To characterize cellular heterogeneity in complex tissues using automated single-cell RNA sequencing workflows.
Materials and Equipment:
- 10x Genomics Chromium Controller or equivalent automated partitioning system
- Fontus NGS liquid handler or equivalent automated workstation
- Single-cell suspension at 700-1,200 cells/μL viability >90%
- Chromium Single Cell 3' Library Kit v3 or equivalent
- Zephyr G3 workstation for automated processing
- Bioanalyzer or TapeStation for quality control
- Illumina NovaSeq 6000 or NextSeq 2000 sequencing platform
Methodology:
- Single-Cell Isolation and Barcoding:
- Prepare single-cell suspension using enzymatic dissociation and filtering.
- Utilize automated systems like the 10x Genomics Chromium to partition individual cells into nanoliter-scale droplets with barcoded beads.
- Automate cell lysis and mRNA capture onto barcoded beads.
Automated Library Preparation:
- Reverse transcription on automated liquid handlers to generate cDNA with cell-specific barcodes.
- cDNA amplification using automated thermal cycling with precise temperature control.
- Library construction through automated fragmentation, end-repair, A-tailing, adapter ligation, and sample cleanup.
- Implement automated size selection using bead-based cleanup methods.
Library Quantification and Quality Control:
- Quantify libraries using qPCR-based methods (NEB Library Quant Kit) for accurate concentration measurement.
- Assess library size distribution using capillary electrophoresis (Agilent Bioanalyzer).
- Normalize libraries to 4nM using automated liquid handlers.
Pooling and Sequencing:
- Pool up to 10 libraries per lane based on calculated molarities.
- Sequence on appropriate Illumina platform with minimum of 20,000 reads per cell.
- Utilize 28bp Read 1 (cell barcode and UMI), 8bp i7 index, and 91bp Read 2 (transcript).
Data Analysis:
- Process raw data through Cell Ranger pipeline for barcode processing, alignment, and counting.
- Perform quality control to remove empty droplets and damaged cells.
- Conduct dimensionality reduction (PCA, UMAP), clustering, and differential expression analysis.
- Utilize trajectory inference and cell-type annotation tools.
Key Automation Advantages:
- Hands-on time reduction from 4 hours to 45 minutes for library preparation [44]
- Elimination of user-to-user variability in repetitive pipetting steps
- Significant reduction in contamination risk through minimal human interaction
- Batch-to-batch consistency across multiple experiments
- Increased throughput with capacity to process 96 samples simultaneously
Single-Cell Analysis Workflow
Diagram 2: Automated single-cell RNA sequencing workflow. Automation significantly improves reproducibility in the barcoding and library preparation steps.
Key Performance Metrics in Automated Single-Cell Analysis
Table 2: Quantitative performance metrics for automated single-cell and spatial NGS workflows
| Parameter | Manual Workflow | Automated Workflow | Improvement |
|---|---|---|---|
| Hands-on Time | 4-6 hours | 45 minutes [44] | >75% reduction |
| Cell Throughput | 1,000-10,000 cells | 10,000-100,000 cells | 10x increase |
| Library Prep Cost | $X | $(X-25%) | 25% reduction |
| Technical Variation | CV 15-25% | CV <5% [47] | >70% improvement |
| Sample Multiplexing | 8-16 samples | 96-384 samples [44] | 6-24x increase |
| Success Rate | 85-90% | 98-99% | >10% improvement |
| Data Consistency | Moderate (user-dependent) | High (standardized) | Significant improvement |
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key research reagents and materials for automated NGS workflows
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Unique Molecular Identifiers (UMIs) | Error correction for PCR and sequencing | Enables accurate quantification and distinguishes biological variants from technical artifacts [48] |
| Barcoded Adapters | Sample multiplexing | Allows pooling of multiple libraries; essential for high-throughput applications [47] |
| Automated Library Prep Kits | Standardized reagent formulations | Optimized for automated liquid handlers; reduces protocol optimization time [44] |
| Size Selection Beads | Fragment size selection | Critical for insert size consistency; automated protocols improve reproducibility [44] |
| qPCR Quantification Kits | Library quantification | Gold standard method; provides accurate concentration for normalization [47] |
| Quality Control Reagents | Assessment of library quality | Includes Bioanalyzer/TapeStation reagents and Qubit assays for DNA/RNA quantification |
| Nuclease-free Water | Dilution and reconstitution | Essential for preventing RNA/DNA degradation in automated systems |
| Enzyme Mixes | Library construction | Includes fragmentation, end-repair, A-tailing, and ligation enzymes in optimized buffers |
Data Analysis Frameworks for Automated NGS
Automated Bioinformatics Pipelines
The massive data volumes generated by automated NGS workflows necessitate robust, automated bioinformatics solutions. Platforms like the CSI NGS Portal provide fully automated NGS data analysis through user-friendly web interfaces, offering 16 standard pipelines for analysing data from DNA, RNA, smallRNA, ChIP, RIP, 4C, SHAPE, circRNA, eCLIP, Bisulfite and scRNA sequencing [49]. These platforms bridge the gap between biologists and bioinformaticians by providing one-click data analysis and sharing capabilities without requiring advanced computational skills [49].
The standard NGS data analysis workflow comprises three core stages:
- Primary Analysis: Quality assessment of raw sequencing data and conversion to FASTQ format, typically performed by software built into the sequencer [48].
- Secondary Analysis: Conversion of data to biological results through alignment, variant calling, and expression quantification using tools like BWA, Bowtie 2, and GATK [48].
- Tertiary Analysis: Biological interpretation and visualization of results to draw conclusions about genetic features, expression patterns, or mutations of interest [48].
NGS Data Analysis Workflow
Diagram 3: Automated NGS data analysis workflow. Platforms like CSI NGS Portal automate these steps from raw data to biological interpretation.
Automated NGS workflows have transformed oncology, infectious disease surveillance, and single-cell analysis by significantly reducing hands-on time, improving reproducibility, and increasing throughput. The integration of robotic liquid handling, standardized reagent kits, and automated bioinformatics pipelines has addressed critical bottlenecks in library preparation and data analysis, enabling researchers to focus on scientific interpretation rather than technical execution. As these technologies continue to evolve, they promise to further accelerate drug discovery and development within the chemogenomics research paradigm, ultimately contributing to more personalized and effective therapeutic interventions.
For research institutions and pharmaceutical companies implementing these workflows, the key considerations include selecting appropriate automation platforms based on projected throughput needs, establishing robust quality control metrics, and investing in bioinformatics infrastructure or partnerships to handle the substantial data generation. With proper implementation, automated NGS workflows provide a powerful foundation for high-throughput chemogenomics research, enabling systematic investigation of compound-biological system interactions at unprecedented scale and resolution.
Optimizing Performance and Overcoming Bottlenecks in Automated NGS Pipelines
In high-throughput chemogenomics research, the reliability of next-generation sequencing (NGS) data is paramount. Automated NGS workflows have revolutionized scalability, but this amplification of throughput also magnifies the impact of any data quality issues, potentially compromising drug discovery pipelines [9]. Real-time quality control (QC) has therefore become a critical component, enabling immediate intervention and ensuring that only high-quality samples progress to downstream analysis. Moving beyond traditional post-experiment QC checks, real-time monitoring integrates quality assessment throughout the automated workflow, preserving valuable reagents, saving time, and safeguarding the integrity of final results [9]. This application note details the implementation of a robust, real-time QC framework specifically for automated NGS workflows in a chemogenomics context.
Key Quality Metrics for Real-Time Monitoring
Effective real-time QC hinges on tracking the right metrics at the right time. The following metrics should be monitored throughout the NGS workflow to assess sample integrity and sequencing performance.
Table 1: Core NGS Quality Metrics for Real-Time Monitoring
| Metric | Description | Target Value/Range | Stage of Assessment |
|---|---|---|---|
| Nucleic Acid Purity (A260/A280) | Assesses protein contamination in DNA/RNA samples [50]. | DNA: ~1.8; RNA: ~2.0 [50] | Nucleic Acid Isolation |
| RNA Integrity Number (RIN) | Quantitative measure of RNA quality [50]. | ≥ 8 for most applications [50] | Nucleic Acid Isolation |
| Library Concentration | Quantifies the yield of the prepared library [8]. | Platform-dependent | Library Preparation |
| Library Fragment Size | Distribution of fragment sizes in the final library [8]. | Platform- and application-dependent | Library Preparation |
| Q Score | Probability of an incorrect base call; Q30 indicates a 1 in 1000 error rate [50]. | ≥ 30 [50] | Sequencing |
| % Bases ≥ Q30 | Percentage of bases with a quality score of 30 or higher [50]. | > 80% | Sequencing |
| Cluster Density | Number of clusters per mm² on the flow cell [50]. | Within optimal range for the sequencer | Sequencing |
| % Clusters Passing Filter (PF) | Percentage of clusters that pass signal purity filters [50]. | > 80% | Sequencing |
| Error Rate | Percentage of bases incorrectly called [50]. | As low as possible, typically < 1% | Sequencing |
| Phasing/Prephasing | Signal loss from clusters falling behind or ahead in sequencing cycles [50]. | < 1% per cycle | Sequencing |
Recent large-scale statistical analyses of public datasets, such as those from the ENCODE project, confirm that while these metrics are fundamental, their relevance and optimal thresholds can vary with specific experimental conditions (e.g., cell type, assay type) [51]. Therefore, establishing condition-specific baselines is a critical best practice.
Implementing a Real-Time QC Workflow
A real-time QC strategy requires integration at every stage of the NGS process. The following workflow diagram and accompanying protocol outline this integrated approach.
Diagram 1: Real-time QC workflow for automated NGS.
Protocol: Real-Time QC Integration in Automated NGS
Objective: To integrate real-time quality checkpoints into an automated NGS workflow for immediate identification and remediation of quality failures.
Materials:
- Automated liquid handling system (e.g., Tecan Fluent, I.DOT Liquid Handler) [52] [53]
- NGS library preparation automation system (e.g., G.PREP NGS Automation solution) [53]
- Automated DNA/RNA quality control instruments (e.g., Agilent TapeStation, Thermo Fisher NanoDrop, fluorometers) [50]
- QC analysis software (e.g., FastQC, NanoPlot, omnomicsQ) [9] [51] [50]
- Laboratory Information Management System (LIMS)
Methodology:
Post-Nucleic Acid Isolation QC:
- Automated Transfer: Upon completion of automated nucleic acid extraction, the system transfers an aliquot of each sample to a dedicated QC analysis station.
- Purity & Integrity Assessment: Automated instruments assess sample purity (A260/A280) and integrity (e.g., RIN via TapeStation) [50].
- Real-Time Decision: Results are automatically parsed by the LIMS or QC software (e.g., omnomicsQ). Samples failing pre-set thresholds are flagged and excluded from downstream library preparation, preventing reagent waste on compromised material [9].
Post-Library Preparation QC:
- Automated Quantification and Sizing: The automated workflow includes a step where the final library is quantified (e.g., via fluorometry or qPCR) and its fragment size distribution is analyzed [8].
- Normalization: Library concentrations are used to automatically calculate and dispense volumes required for equimolar pooling, ensuring balanced sequencing representation.
- Quality Gate: Libraries with insufficient concentration, incorrect average size, or abnormal size distribution are flagged and do not proceed to sequencing.
In-Run Sequencing QC:
- Live Monitoring: During the sequencing run, key performance metrics—including Q scores, cluster density, % PF, phasing/prephasing, and error rate—are monitored in real-time via the sequencing instrument's software [50].
- Automated Alerts: The system is configured to trigger alerts if any metric deviates significantly from expected values, allowing for potential early termination of a failing run and conserving sequencing capacity.
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful implementation of this protocol relies on specific reagents and tools. The following table details essential solutions for ensuring data quality in automated NGS workflows.
Table 2: Key Research Reagent Solutions for NGS QC
| Item | Function | Example Use Case |
|---|---|---|
| Automated NGS Clean-Up Kits | Magnetic bead-based purification of nucleic acids during library prep; removes enzymes, salts, and short fragments [53]. | Integrated into automated liquid handling protocols for consistent post-enzymatic reaction clean-up. |
| NGS Library Prep Kits | Reagent kits optimized for automation, providing pre-mixed, stable reagents for robust performance with minimal hands-on time. | Used on automated platforms for highly reproducible DNA or RNA library construction. |
| QC Assay Kits | Kits for fluorometric quantification (e.g., dsDNA HS Assay) or qPCR-based library quantification. | Automated quantification of nucleic acids and final libraries prior to sequencing. |
| Automated Liquid Handlers | Robots that precisely dispense sub-microliter volumes of samples and reagents, eliminating pipetting variability and cross-contamination [9] [53]. | Performing all liquid transfer steps in nucleic acid extraction, library prep, and QC setup. |
| Real-Time QC Software | Software tools (e.g., omnomicsQ, FastQC) that automatically analyze QC data, compare to thresholds, and flag anomalies [9] [50]. | Providing the decision-making engine for real-time quality gates throughout the workflow. |
Data Analysis and Interpretation
The final QC checkpoint involves analyzing the raw sequencing data. Tools like FastQC provide a comprehensive overview of read quality, per-base sequence content, adapter contamination, and other potential issues [50]. For real-time assessment, these tools can be run on a subset of initial sequencing data to predict the overall success of the run.
As emphasized by large-scale studies, the interpretation of QC data should be condition-specific. For instance, the ENCODE guidelines for uniquely mapped reads may not reliably differentiate between high- and low-quality files in all assay types [51]. Therefore, leveraging data-driven, condition-specific statistical guidelines is recommended for accurate quality classification.
Implementing a robust real-time QC and monitoring system is not an optional enhancement but a core requirement for automated, high-throughput chemogenomics. By integrating the metrics, protocols, and tools outlined in this document, research and drug development teams can achieve a new level of operational efficiency and data reliability. This proactive approach to quality management minimizes costly repeats, accelerates discovery timelines, and ensures that critical decisions are based on the highest quality genomic data.
Next-Generation Sequencing (NGS) has become a foundational technology in high-throughput chemogenomics research, enabling unprecedented insights into genetic variations, gene expression, and drug mechanisms. However, the scale and complexity of data generated present substantial computational challenges that can bottleneck research progress. Effectively managing terabytes of sequencing data requires integrated strategies spanning specialized hardware, optimized software, cloud computing infrastructure, and automated workflows. This application note details practical strategies and protocols for overcoming these computational barriers, with a specific focus on their application within automated chemogenomics research. We provide benchmarked methodologies to help researchers and drug development professionals maintain analytical rigor while scaling their genomic investigations.
Computational Framework and Core Components
Integrated Data Analysis Strategy
A robust computational framework for large-scale NGS data integrates high-performance computing resources with specialized analytical pipelines. This synergy is crucial for transforming raw sequencing data into biologically interpretable results, particularly in high-throughput chemogenomics where analyzing chemical-genetic interactions systematically is paramount. The core strategy involves leveraging cloud computing for scalable infrastructure, specialized pipelines for accelerated processing, and automated workflows to ensure reproducibility and efficiency [54] [55] [52].
The diagram below illustrates the integrated computational framework that connects these components from sample to insight in chemogenomics research.
Research Reagent Solutions and Essential Materials
Successful implementation of computational strategies requires specific reagent solutions and analytical tools. The following table details key components essential for automated NGS workflows in chemogenomics research.
Table 1: Essential Research Reagent Solutions for Automated NGS Workflows
| Solution/Material | Function in Workflow | Application Context |
|---|---|---|
| IDT xGen NGS Products [56] | Hybrid capture probes for target enrichment | Customizable target sequencing for cancer research and biomarker discovery |
| Archer FUSIONPlex/VARIANTPlex [57] | Targeted RNA/DNA sequencing assays | Fusion gene detection and variant screening in oncology |
| Hamilton Microlab STAR [56] | Automated liquid handling system | High-throughput reagent dispensing and library preparation automation |
| Biomek i3 Benchtop Liquid Handler [57] | Compact liquid handling workstation | Automated NGS library prep for low-to-mid throughput labs |
| Illumina Connected Analytics [58] | Cloud-based genomic data platform | Multi-omic data management, analysis, and sharing |
| Sentieon DNASeq [55] | Accelerated variant calling pipeline | Rapid germline variant analysis for clinical applications |
| NVIDIA Clara Parabricks [55] | GPU-accelerated variant calling | Ultra-rapid secondary analysis of WGS/WES data |
| DeepCE [59] | Deep learning for gene expression prediction | Predicting chemical-induced gene expression profiles for drug repurposing |
Protocols for High-Performance NGS Data Analysis
Protocol: Cloud-Based Deployment of Ultra-Rapid NGS Pipelines
Objective: To deploy and execute accelerated germline variant calling pipelines on Google Cloud Platform (GCP) for rapid turnaround of whole genome (WGS) and whole exome (WES) data.
Background: Sentieon DNASeq and Clara Parabricks Germline represent state-of-the-art solutions for secondary NGS analysis, significantly reducing computation time from days to hours. Sentieon optimizes CPU utilization, while Parabricks leverages GPU acceleration [55].
Materials:
- Raw sequencing data in FASTQ format
- GCP account with billing enabled
- Reference genome (GRCh38) and associated index files
- Sentieon software license OR Clara Parabricks installation
Methodology:
- Virtual Machine (VM) Configuration:
Data Transfer:
- Upload FASTQ files to Google Cloud Storage bucket.
- Download reference files (genome, dbsnp, known indel sites) to VM.
Pipeline Execution - Sentieon DNASeq:
Pipeline Execution - Clara Parabricks:
Output and Cleanup:
- Download VCF results to local storage.
- Stop or delete VM instances to minimize costs.
- Archive input data in cold storage (e.g., Nearline Storage) if needed.
Technical Notes:
- Always validate pipeline setup with a small test dataset before processing full batches.
- Configure cost alerts in GCP to monitor and control expenditure.
- For clinical data, ensure VMs comply with HIPAA and GDPR requirements through GCP's compliance features [55] [9].
Protocol: AI-Enhanced Prediction of Chemical-Induced Gene Expression
Objective: To predict genome-wide gene expression profiles induced by novel chemical compounds using the DeepCE deep learning framework, enabling mechanism-driven phenotype screening for drug repurposing.
Background: DeepCE utilizes graph neural networks and multi-head attention mechanisms to model chemical substructure-gene and gene-gene associations, predicting differential gene expression for de novo chemicals without requiring physical screening [59].
Materials:
- Chemical structures in SMILES format
- L1000 reference dataset (Bayesian-based peak deconvolution version)
- DeepCE software (publicly available)
- STRING database for protein-protein interaction networks
- High-performance computing environment with GPU support
Methodology:
- Data Preparation:
- Standardize chemical structures and remove duplicates.
- Encode molecular structures as graphs for graph convolutional network input.
Model Configuration:
- Implement DeepCE architecture with graph neural network (GCN) for substructure feature extraction.
- Configure multi-head attention layers to capture chemical-gene associations.
- Set up multi-output, multilayer feed-forward network for gene expression prediction.
Training and Validation:
- Split data using time-series cross-validation to prevent data leakage.
- Utilize data augmentation to extract information from unreliable L1000 experiments.
- Validate model performance using root mean square error (RMSE) and correlation coefficients.
Expression Prediction:
Downstream Application:
- Compare predicted profiles with disease signatures for drug repurposing.
- Identify potential mechanism of action through enriched pathways.
- Prioritize candidate compounds for experimental validation.
Technical Notes:
- The DeepCE framework has been successfully applied to COVID-19 drug repurposing, identifying compounds consistent with clinical evidence [59].
- Predictions are most reliable for cell lines and dosage ranges well-represented in the training data.
- Always corroborate in silico predictions with targeted experimental validation.
Experimental Results and Benchmarking Data
Implementation of the described strategies yields significant improvements in processing efficiency and cost management. The table below summarizes benchmark data for cloud-based pipeline execution.
Table 2: Performance Benchmarking of Ultra-Rapid NGS Pipelines on GCP
| Pipeline | Sample Type | Average Runtime (Hours) | Cost per Sample ($) | CPU/GPU Utilization | Optimal Use Case |
|---|---|---|---|---|---|
| Sentieon DNASeq [55] | WGS | 2.1 | 3.76 | 98% CPU | High-throughput clinical sequencing |
| Sentieon DNASeq [55] | WES | 0.8 | 1.43 | 95% CPU | Targeted sequencing studies |
| Clara Parabricks [55] | WGS | 1.7 | 2.81 | 92% GPU | Rapid-turnaround diagnostics |
| Clara Parabricks [55] | WES | 0.6 | 0.99 | 88% GPU | Research requiring fastest results |
| Basepair SaaS [60] | Varied | Variable | Pay-per-sample | Managed service | Labs seeking minimal IT overhead |
Implementation Guide for Automated Chemogenomics Workflows
Strategic Integration of Automated NGS Platforms
Successful implementation of automated NGS workflows in chemogenomics requires careful planning and cross-disciplinary collaboration. The diagram below outlines the complete automated workflow from experimental design to biological insight, highlighting critical decision points.
Critical Implementation Considerations
Workflow Integration: Ensure automated liquid handling systems (e.g., Hamilton Microlab STAR, Biomek i3) seamlessly integrate with laboratory information management systems (LIMS) and downstream analysis platforms [56] [9] [57].
Data Security and Compliance: For clinical chemogenomics applications, implement solutions that comply with HIPAA, GDPR, and IVDR regulations through encryption, access controls, and audit trails [54] [9].
Cost Management: Leverage cloud cost management tools and storage tiering (e.g., Basepair's automated archival) to reduce expenses by up to 80% compared to on-premises solutions [55] [60].
Personnel Training: Develop comprehensive training programs covering automated system operation, workflow software, troubleshooting, and regulatory requirements to ensure smooth technology adoption [9] [52].
The strategies and protocols outlined provide a comprehensive framework for managing computational challenges in large-scale NGS data analysis. By implementing these integrated approaches, research organizations can significantly accelerate chemogenomics discovery while maintaining analytical rigor and cost-effectiveness.
The rapid integration of next-generation sequencing (NGS) into high-throughput chemogenomics research has fundamentally transformed drug discovery and development. This growth is substantiated by market projections estimating the NGS market will expand from $12.13 billion in 2023 to approximately $23.55 billion by 2029, reflecting a compound annual growth rate of about 13.2% [35]. However, the full potential of automated NGS in generating reproducible, high-quality data can only be realized through rigorous standardization protocols that address the entire workflow from sample preparation to data analysis. The inherent complexity of NGS technologies, coupled with the demanding precision required for chemogenomics applications, necessitates a systematic approach to quality management that ensures reliable and consistent results across experiments, platforms, and laboratories [61] [62].
Standardization in this context extends beyond mere protocol consistency; it encompasses the comprehensive implementation of Quality System Essentials (QSE) that govern personnel competency, equipment management, process control, and data handling. The Centers for Disease Control and Prevention (CDC) and Association of Public Health Laboratories (APHL) collaboration through the Next-Generation Sequencing Quality Initiative (NGS QI) addresses these exact challenges by developing tools and resources specifically designed to build robust quality management systems for NGS workflows [61]. Similarly, the American College of Medical Genetics and Genomics (ACMG) has established clinical laboratory standards for NGS to ensure consistency in clinical applications, providing a valuable framework for research settings [63]. For chemogenomics researchers, these standardized approaches are indispensable for generating reproducible data that accurately elucidates compound-genome interactions, enables reliable biomarker discovery, and supports valid therapeutic predictions.
Critical Challenges in NGS Standardization
Technical and Workflow Variability
The path to standardized NGS workflows is fraught with technical challenges that introduce variability and compromise reproducibility. A primary concern is the library preparation complexity, where manual handling and pipetting introduce significant variability in reagent dispensing, incubation times, and sample tracking [9]. This variability directly impacts sequencing accuracy and consistency, particularly in high-throughput chemogenomics screens where uniform library quality is paramount for comparing compound effects across thousands of genetic targets. The problem is exacerbated by the diversity of available NGS platforms, each with unique chemistries, template preparation methods (clonally amplified, single-molecule, or circle templates), and sequencing-by-synthesis approaches (cyclic reversible termination, sequencing by ligation, single-nucleotide addition via pyrosequencing, and real-time sequencing) that yield different read lengths, accuracy profiles, and error rates [64].
Furthermore, the rapid technological evolution in NGS presents a persistent standardization challenge. As noted by the NGS QI, new kit chemistries from Oxford Nanopore Technologies that utilize CRISPR for targeted sequencing and improved basecaller algorithms employing artificial intelligence and machine learning continuously emerge, enhancing accuracy but requiring frequent revalidation of established workflows [61]. Similarly, emerging platforms like Element Biosciences demonstrate increasing accuracies at lower costs, encouraging migration from older systems but necessitating complete revalidation [61]. This dynamic technological landscape creates a tension between adopting improvements and maintaining standardized, validated workflows, particularly for regulated drug development environments where consistency is paramount.
Personnel and Data Management Hurdles
The human element represents another critical challenge in NGS standardization. Workforce retention of proficient personnel poses a substantial obstacle due to the unique and specialized knowledge required, which in turn increases costs for adequate staff compensation [61]. Akkari et al. found that some testing personnel held their positions for <4 years on average, and in 2021, APHL reported that 30% of surveyed public health laboratory staff indicated an intent to leave the workforce within the next 5 years [61]. This turnover directly threatens protocol consistency and requires robust documentation and training systems to mitigate.
The bioinformatic analysis phase introduces additional standardization hurdles, often described as a "next-generation gap" between data generation and analytical interpretation [64]. The absence of uniform data formats, processing pipelines, and variant calling algorithms compromises result comparability across studies and laboratories. This challenge is particularly acute in chemogenomics, where integrating NGS data with chemical compound information demands rigorous computational standardization to ensure valid structure-activity relationship determinations. Data heterogeneity, model interpretability, and ethical concerns further complicate the implementation of standardized AI and machine learning approaches for NGS analysis [52].
Table 1: Key Challenges in NGS Standardization and Their Impacts on Chemogenomics Research
| Challenge Category | Specific Challenges | Impact on Chemogenomics Research |
|---|---|---|
| Technical Variability | Library preparation inconsistencies; Platform diversity; Rapid technological evolution | Reduced reproducibility of compound-genome interaction studies; Inconsistent biomarker identification; Impaired cross-study comparisons |
| Personnel Factors | Specialized staff turnover; Training requirements; CLIA regulations for qualified personnel | Protocol deviations; Increased validation costs; Extended implementation timelines for automated systems |
| Data Management | Bioinformatics pipeline variability; Non-standardized data formats; AI/ML integration complexity | Inconsistent variant calling; Challenges integrating chemical and genomic data; Limited dataset reusability |
| Quality Systems | Lack of standardized SOPs; Variable quality metrics; Regulatory compliance burden | Increased false positives/negatives in compound screening; Difficult technology transfer; Barriers to regulatory approval |
Standardized Framework for Automated NGS Workflows
Quality Management System Essentials
Implementing a comprehensive Quality Management System (QMS) forms the foundation for standardized and reproducible automated NGS workflows. The NGS QI emphasizes that a robust QMS enables continual improvement and proper document management in laboratories, with all products undergoing review every three years to ensure they remain current with technology, standard practices, and regulatory changes [61]. For chemogenomics research, this translates to developing standardized protocols that address the entire workflow while maintaining flexibility to accommodate specific research objectives and technology platforms.
The core components of an effective QMS for automated NGS include standard operating procedures (SOPs) for all critical processes, equipment management protocols with regular calibration and maintenance schedules, personnel competency assessment programs, and document control systems that manage protocol versions and revisions. Particularly valuable are the resources developed by the NGS QI, including the QMS Assessment Tool, Identifying and Monitoring NGS Key Performance Indicators SOP, NGS Method Validation Plan, and NGS Method Validation SOP, which provide templates that laboratories can adapt to their specific automated workflows [61]. These tools help establish the systematic approach necessary for generating consistent, reproducible results in high-throughput chemogenomics applications.
Cross-laboratory standardization is further enhanced through participation in External Quality Assessment (EQA) programs, such as those organized by EMQN and GenQA, which help laboratories benchmark their workflows against industry standards [9]. Additionally, compliance with quality standards like ISO 13485 and adherence to guidelines from professional organizations like ACMG and College of American Pathologists (CAP) provide structured frameworks for quality assurance [63] [9]. For drug development professionals, implementing these standardized quality systems not only improves data reliability but also facilitates regulatory submissions by demonstrating rigorous process control.
Automated Workflow Implementation
The integration of automation technologies represents a pivotal strategy for achieving standardization in NGS workflows. Automated systems address key sources of variability by ensuring precise reagent dispensing, reducing cross-contamination risks through disposable tips and controlled aspiration speeds, and enforcing strict adherence to validated protocols [9]. The benefits are quantifiable: a recent study at Heidelberg University Hospital demonstrated that automating NGS workflows reduced manual hands-on time from approximately 23 hours per run to just six hours – a nearly four-fold decrease – while simultaneously improving key performance metrics, including a higher percentage of aligned reads (increasing from approximately 85% to 90%) [35].
Strategic implementation of automated NGS workflows requires careful consideration of several factors. First, laboratories must assess their specific needs, including sample volume, required throughput, and regulatory requirements [9]. For high-throughput chemogenomics research processing hundreds or thousands of compound samples, automation platforms with high scalability and integration capabilities with existing Laboratory Information Management Systems (LIMS) are essential. Second, selecting the appropriate automation platform requires verifying compatibility with current NGS pipelines, bioinformatics tools, and regulatory frameworks to prevent disruptions and ensure data integrity [9]. Third, personnel must receive comprehensive training on new protocols, software, and compliance requirements to ensure smooth operational transition and maximize the benefits of automation.
Table 2: Quantitative Benefits of NGS Automation Demonstrated in Comparative Studies
| Performance Metric | Manual Process | Automated Process | Improvement |
|---|---|---|---|
| Hands-on Time (per run) | ~23 hours [35] | ~6 hours [35] | 73% reduction |
| Total Runtime | 42.5 hours [35] | 24 hours [35] | 44% reduction |
| Aligned Reads | ~85% [35] | ~90% [35] | 5 percentage point increase |
| Library Yield | 2.4 pmol (manual) [35] | 3.1 pmol (automated) [35] | 29% increase |
| On-target Rate | <90% (manual) [35] | >90% (automated) [35] | Significant improvement |
| Cross-contamination Risk | High [9] | Minimal [9] | Substantial reduction |
Standardized Automated NGS Workflow with Quality Gates
Experimental Protocols for Standardized Automated NGS
Automated Library Preparation Protocol
Principle: This protocol standardizes the library preparation phase of NGS workflows using automated liquid handling systems to minimize variability, increase reproducibility, and ensure consistent library quality for chemogenomics applications.
Materials:
- Automated liquid handling system (e.g., Beckman Coulter Biomek i7, Tecan Fluent)
- Library preparation kit (e.g., Illumina Nextera Flex, Twist NGS)
- DNA/RNA samples quantified by fluorometry
- Low-binding microplates and tips
- Magnetic bead-based purification kit
- Real-time quality control system (e.g., omnomicsQ)
Procedure:
- Sample Quality Control: Confirm sample concentration and purity using fluorometric methods. DNA samples should have A260/A280 ratio of 1.8-2.0 and minimum concentration of 15 ng/μL for amplified templates or <1 μg for single-molecule templates [64].
- System Calibration: Verify automated liquid handler calibration using dye-based volume verification tests. Ensure all reagents are properly thawed, mixed, and centrifuged before loading onto the system.
- Normalization and Transfer: Program the automated system to normalize all samples to the required concentration and transfer equal volumes to the reaction plate.
- Fragmentation and End-Repair: Initiate automated fragmentation program following manufacturer specifications. For enzymatic fragmentation, ensure precise temperature control during incubation steps.
- Adapter Ligation: Program system to add unique dual-indexed adapters to each sample using precise liquid handling to minimize cross-contamination. Incubate according to manufacturer recommendations.
- Library Amplification: Perform PCR amplification with cycle number optimized for input material. Automated systems should monitor temperature uniformity across all samples.
- Purification and Normalization: Execute magnetic bead-based clean-up with precisely controlled incubation times and mixing parameters. Normalize final libraries to equimolar concentrations.
- Quality Control: Transfer aliquot of each library to quality control analysis, including fragment size distribution (e.g., TapeStation, Bioanalyzer) and quantification (qPCR).
Quality Control Parameters:
- Library concentration: ≥2 nM
- Fragment size distribution: within expected range for application (e.g., 300-500 bp for whole genome sequencing)
- Adapter dimer: ≤5% of total fragments
- Percentage of aligned reads: ≥90% [35]
NGS Method Validation Protocol
Principle: Establish performance characteristics of automated NGS workflows to ensure reliability, reproducibility, and accuracy for chemogenomics research applications, following guidelines from ACMG, CAP, and NGS QI resources.
Materials:
- Reference materials with known variants (e.g., Genome in a Bottle, Horizon Multiplex)
- Clinical or research samples with established genotypes
- All reagents and consumables for automated NGS workflow
- Bioinformatics pipeline with version-controlled components
Procedure:
- Validation Planning: Define validation scope, including platform, test method, and intended use. Establish acceptance criteria for accuracy, precision, sensitivity, specificity, and reproducibility based on ACMG standards [63].
- Sample Selection: Include reference materials with variants across different genomic contexts (SNVs, indels, CNVs), spanning a range of allele frequencies (5-95%). Include at least 30 positive and 30 negative samples for each variant type.
- Experimental Runs: Perform multiple independent runs (minimum of 3) on different days with different operators to assess inter-run variability. Include replicates within each run to assess intra-run variability.
- Data Analysis: Process data through standardized bioinformatics pipeline. Compare variant calls to expected results from reference materials.
- Performance Calculation: Calculate accuracy, precision, sensitivity, and specificity using established formulas:
- Sensitivity = True Positives / (True Positives + False Negatives)
- Specificity = True Negatives / (True Negatives + False Positives)
- Precision = True Positives / (True Positives + False Positives)
Acceptance Criteria:
- Sensitivity: ≥99% for SNVs and indels at ≥5% allele frequency
- Specificity: ≥99% for all variant types
- Reproducibility: ≥95% concordance between replicates and runs
- Coverage uniformity: ≥90% of targets at ≥100x coverage
Research Reagent Solutions for Standardized NGS
Table 3: Essential Research Reagents and Their Functions in Automated NGS Workflows
| Reagent Category | Specific Examples | Function in NGS Workflow | Standardization Considerations |
|---|---|---|---|
| Library Prep Kits | Illumina Nextera Flex; Twist NGS; Pillar Biosciences panels | Fragment DNA, add adapters, amplify libraries | Kit lot tracking; Protocol optimization for automation; Validation against reference materials |
| Enzymes & Master Mixes | Watchmaker Genomics enzymes; High-fidelity polymerases | Catalyze fragmentation, end-repair, ligation, amplification | Activity verification; Storage condition monitoring; Stability testing |
| Normalization Beads | SPRIselect; AMPure XP | Size selection and purification of libraries | Lot-to-lot performance validation; Volume calibration for automated systems |
| Quality Control Assays | Qubit dsDNA HS; TapeStation D1000; Fragment Analyzer | Quantify and qualify input DNA and final libraries | Regular calibration; Inclusion of standards; Threshold establishment |
| Reference Materials | Genome in a Bottle; Horizon Multiplex controls | Process monitoring; Assay validation; Quality assurance | Proper storage; Aliquot management; Documentation of usage |
| Index Adapters | Illumina Dual Indexes; IDT for Illumina | Sample multiplexing; Library identification | Unique dual-index implementation; Index balancing; Cross-contamination monitoring |
Quality Control and Data Management
Quality Monitoring Systems
Implementing robust quality monitoring systems throughout the automated NGS workflow is essential for maintaining standardization and ensuring reproducible results. Real-time quality control tools, such as omnomicsQ, provide continuous assessment of sample quality, allowing detection of issues before they compromise downstream analysis [9]. These systems flag samples that fall below pre-defined quality thresholds, preventing low-quality samples from advancing in the workflow and conserving valuable reagents and sequencing resources.
The NGS QI emphasizes the importance of identifying and monitoring Key Performance Indicators (KPIs) as part of a comprehensive quality management system [61]. For automated NGS workflows in chemogenomics research, critical KPIs include:
- Library preparation efficiency: Conversion rate of input DNA to sequencing-ready library
- Sequence quality metrics: Q-scores, percent bases ≥Q30
- Mapping metrics: Percentage of aligned reads, uniformity of coverage
- Variant calling accuracy: Sensitivity, specificity, and reproducibility for known variants
- Sample cross-contamination: Percentage of reads aligning to incorrect samples
These metrics should be tracked longitudinally using statistical process control methods to identify trends, detect deviations from established baselines, and trigger corrective actions when necessary. The NGS QI's "Identifying and Monitoring NGS Key Performance Indicators SOP" provides a standardized framework for this monitoring process [61].
Bioinformatics Standardization
Standardization of bioinformatic analyses is equally critical for reproducible NGS results in chemogenomics research. This includes implementing version-controlled pipelines, standardized data formats, and consistent variant calling approaches. The integration of artificial intelligence and machine learning in bioinformatics tools, such as DeepVariant for variant calling, has demonstrated improved accuracy compared to traditional heuristic-based approaches but requires careful standardization to ensure consistent performance [52].
A standardized bioinformatics workflow should include:
- Raw data quality control: FastQC or similar tools with standardized parameters
- Adapter trimming and quality filtering: Consistent cut-off values across all samples
- Alignment to reference genome: Standardized reference genome version and alignment parameters
- Post-alignment processing: Duplicate marking, base quality recalibration with standardized parameters
- Variant calling: Consistent algorithms and filtering criteria
- Variant annotation: Standardized annotation sources and versions
Documentation of all software versions, parameters, and reference databases is essential for reproducibility. The use of containerization technologies (Docker, Singularity) and workflow management systems (Nextflow, Snakemake) further enhances reproducibility by encapsulating the complete computational environment.
Quality Management Cycle for NGS Standardization
Standardization and reproducibility in automated NGS workflows are not merely desirable attributes but fundamental requirements for robust chemogenomics research and drug development. The implementation of comprehensive quality management systems, strategic automation integration, standardized experimental protocols, and rigorous bioinformatic pipelines collectively address the critical challenges of variability and inconsistency in NGS applications. As the field continues to evolve with emerging technologies such as AI-enhanced basecalling and third-generation sequencing, the commitment to standardization principles will ensure that these advancements translate into reliable, reproducible results rather than additional sources of variability.
For researchers, scientists, and drug development professionals, embracing these standardized approaches requires initial investment in system development, validation, and personnel training, but yields substantial returns through enhanced data quality, reduced rework, and accelerated discovery timelines. The frameworks and resources provided by organizations such as the NGS Quality Initiative, ACMG, and CAP offer validated starting points for developing laboratory-specific standardization protocols. By maintaining this focus on standardization and reproducibility, the chemogenomics research community can fully leverage the transformative potential of automated NGS workflows to advance therapeutic discovery and precision medicine.
The integration of fully automated next-generation sequencing (NGS) library preparation platforms has transitioned from a niche novelty to an indispensable tool in high-throughput chemogenomics research [65]. These systems, which integrate precise reagent handling, temperature control, and workflow standardization, address critical challenges related to reproducibility and throughput that have long constrained manual protocols [65]. However, the sophistication of these platforms creates a foundational dependency on a highly skilled workforce. The precision and efficiency of automated NGS are not inherent to the machinery alone but are a direct function of the personnel's expertise in its operation, maintenance, and troubleshooting. This document outlines a comprehensive framework for building and sustaining this critical expertise, ensuring that automated systems fulfill their potential to accelerate genomic discovery and improve patient outcomes in chemogenomics research [65].
Core Competency Framework for Automated NGS Operations
Effective personnel training must be structured around a clear set of core competencies. These competencies span technical, analytical, and regulatory domains, ensuring a holistic understanding of the automated NGS workflow.
Table 1: Core Competency Framework for Automated NGS Personnel
| Competency Domain | Key Skills and Knowledge Areas | Importance in Chemogenomics |
|---|---|---|
| System Operation | Automated liquid handling, robotic operation, workflow software configuration, routine start-up/shutdown [9]. | Ensures precise dispensing of chemogenomic libraries and reagents, maintaining assay consistency for high-throughput drug screening. |
| Process Standardization | Adherence to Standard Operating Procedures (SOPs), protocol customization, understanding of enzymatic fragmentation and magnetic bead-based purification chemistries [9] [65]. | Eliminates batch-to-batch variation, which is critical for reproducible compound profiling and biomarker discovery. |
| Quality Control & Monitoring | Operation of QC instruments (e.g., Agilent TapeStation, Thermo Scientific NanoDrop), interpretation of metrics (e.g., RIN, Q scores, adapter content), real-time quality monitoring using tools like omnomicsQ [9] [50]. | Flags low-quality samples early, preventing wasted resources on failed sequencing runs and ensuring data integrity for downstream analysis. |
| Bioinformatics Fundamentals | Understanding of FASTQ format, quality score (Q score) interpretation, and basic principles of read alignment and variant calling [66] [50]. | Enables effective cross-disciplinary communication and preliminary assessment of sequencing run success before deep bioinformatic analysis. |
| Regulatory Compliance & Data Security | Knowledge of IVDR, ISO 13485, ACMG/CAP guidelines, and data protection standards like GDPR and HIPAA [9]. | Essential for labs involved in diagnostic discovery and for maintaining the security of sensitive patient-derived chemogenomic data. |
| Troubleshooting & Maintenance | Ability to identify and resolve common errors (e.g., low PF %, pipetting inconsistencies), perform routine maintenance, and manage supply chains [9] [65]. | Minimizes system downtime, ensuring continuous operation in high-throughput research environments. |
Detailed Experimental Protocol: A Training Exercise for Automated QC and Library Preparation
This hands-on protocol is designed to train personnel in the critical tasks of sample quality control and automated library preparation, emphasizing the points where technique and judgment impact downstream outcomes.
Training Objective
To proficiently execute and quality-control an automated NGS library preparation run for a set of genomic DNA samples, using a defined chemogenomics panel.
Research Reagent Solutions and Essential Materials
Table 2: Key Research Reagent Solutions for Automated NGS Library Prep
| Item | Function | Example & Notes |
|---|---|---|
| NGS Library Prep Kit | Provides all enzymes and buffers for end-repair, A-tailing, and adapter ligation. | Select kits pre-validated for your automation platform [9]. |
| Magnetic Beads | For size selection and purification of the library post-enzymatic steps. | Magnetic bead-based purification is experiencing rapid uptake for its streamlined process [65]. |
| Adapter Oligos | Attach to DNA fragments to enable binding to the sequencing flow cell and sample indexing. | Ensure adapter indices are unique and compatible with your sequencing platform. |
| PCR Master Mix | Amplifies the adapter-ligated DNA library to generate sufficient material for sequencing. | |
| Ethanol (80%) | Used in wash steps with magnetic beads to purify the library. | Must be freshly prepared. |
| Nuclease-Free Water | The elution buffer for the final purified library. |
Methodology
Pre-Run: Quality Control of Input DNA
- Quantification and Purity Check: Using the NanoDrop spectrophotometer, load 1 µL of each genomic DNA sample.
- Action: Record the concentration (ng/µL) and the A260/A280 ratio.
- Acceptance Criteria: A260/A280 ratio of ~1.8 [50]. Deviations suggest protein or other contamination.
- Trainee Decision Point: Should a sample with an A260/A280 ratio of 1.6 be processed? (Answer: No, it should be re-purified).
- Integrity and Size Distribution Check: Using the Agilent TapeStation, analyze 1 µL of each sample that passed step 1.
- Action: Review the electrophoretogram and the assigned DNA Integrity Number (DIN) or equivalent.
- Acceptance Criteria: A clear, high-molecular-weight peak with a DIN >7.0.
- Trainee Decision Point: A sample showing significant smearing below the main peak indicates degradation. Should it proceed? (Answer: No).
Run: Automated Library Preparation
- System Startup and Prime: Power on the automated liquid handling system and associated robotic modules. Execute any required priming or flushing routines for the fluidics system as per the manufacturer's SOP.
- Workflow Software Configuration:
- Log in to the workflow software and select the appropriate NGS library preparation protocol.
- Input the sample manifest, including sample IDs and pre-determined concentrations from the QC step.
- The software will calculate required reagent volumes. The trainee must verify these calculations manually for at least two samples as a cross-check.
- Plate Setup and Reagent Deck Layout:
- On a chilled microtiter plate, load the required volume of each quantified DNA sample into the assigned wells.
- Following the deck layout map, place all reagents (see Table 2) in their designated positions on the deck. Ensure volumes are sufficient for the entire run plus a dead-volume contingency.
- Run Initiation and Monitoring:
- Start the automated run. The system will perform enzymatic fragmentation, end-repair, A-tailing, adapter ligation, and PCR amplification.
- Monitor the run in real-time via the software dashboard for any immediate errors (e.g., liquid level detection failures, clogged tips).
- Post-Run Clean-Up: The system will transfer the final PCR product to a new plate for magnetic bead-based purification. Some systems integrate this step; others require a separate, automated clean-up protocol.
Post-Run: Quality Control of Final Library
- Quantification and Size Analysis: Use the TapeStation to analyze 1 µL of the final, purified library from each sample.
- Action: Confirm the library concentration and inspect the size distribution profile. The profile should show a tight, specific size range without adapter dimer (~120-150 bp).
- Trainee Decision Point: A sample shows a large peak at ~120 bp. What is this likely to be, and can the library be sequenced? (Answer: Adapter dimer; it should be re-purified with adjusted bead ratios to remove it).
- Pooling and Normalization: Based on the QC results, calculate the volumes required to pool libraries at equimolar concentrations for sequencing.
Data Interpretation and Analysis
Trainees should generate a final report summarizing the QC metrics for both input DNA and the final libraries, justifying any decisions to exclude samples and evaluating the overall success of the automated run.
Personnel Competency Assessment and Certification Workflow
A structured pathway from novice to certified operator ensures personnel are fully qualified to operate and maintain automated NGS systems independently. The following diagram visualizes this workflow and the continuous learning cycle.
Strategic Implementation of Training Programs
Implementing a successful training program requires strategic planning beyond the technical protocols. Key considerations include:
- Integrating with Existing Systems: Training must emphasize how the automated platform integrates with the laboratory's Laboratory Information Management System (LIMS) and downstream NGS pipelines and bioinformatics tools to avoid workflow disruptions and ensure data integrity [9] [65].
- Evaluating Return on Investment (ROI): While the initial investment in automation and training might seem considerable, the long-term ROI is justified through reduced reagent waste (via precise dispensing), increased throughput, and a reduction in errors and rework, which lowers operational expenditures [9] [65].
- Fostering a Culture of Continuous Improvement: Encourage certified personnel to participate in External Quality Assessment (EQA) programs (e.g., EMQN, GenQA) and stay updated with new chemistries, software upgrades, and evolving regulatory requirements (e.g., IVDR, ISO 13485) [9]. This transforms the training program from a one-time event into a pillar of ongoing operational excellence.
By adopting this comprehensive framework for personnel training and competency assessment, chemogenomics research laboratories can build the expertise necessary to fully leverage automated NGS systems, thereby ensuring the generation of high-quality, reproducible data essential for accelerating drug discovery and development.
Benchmarking Success: Validating Automated NGS Workflows for Robust Drug Discovery
Within high-throughput chemogenomics research, the demand for rapid and reliable genetic data is paramount for accelerating drug discovery and personalized treatment strategies. Next-Generation Sequencing (NGS) serves as a foundational technology in this field, yet its utility hinges on the efficiency and accuracy of the library preparation workflow. This application note provides a detailed, data-driven comparison between manual and automated NGS workflows, focusing on three critical performance metrics: turnaround time (TAT), hands-on time, and error rates. The objective quantification of these metrics is essential for laboratories aiming to scale their operations, enhance reproducibility, and integrate NGS seamlessly into high-throughput chemogenomics pipelines.
Quantitative Performance Metrics
Data from controlled studies and real-world implementations consistently demonstrate the superior performance of automated NGS workflows. The table below summarizes key quantitative comparisons between automated and manual methods.
Table 1: Comparative Performance Metrics of Automated vs. Manual NGS Workflows
| Performance Metric | Manual Workflow | Automated Workflow | Key Findings and Context |
|---|---|---|---|
| Overall Turnaround Time (TAT) | Typically multiple days [67] | ~24 hours from sample to result [68] | Automated, integrated systems significantly reduce total TAT, enabling faster clinical decision-making. [68] |
| Hands-On Time for Nucleic Acid Extraction | ~120 minutes [68] | ~30 minutes [68] | Automation reduces active technologist time by 75% for this initial step, freeing personnel for other tasks. [68] |
| Library Prep Hands-On Time | High; several hours [69] | 65% or greater reduction [18] | Automated liquid handling slashes manual input; some systems report over 65% less hands-on time. [18] |
| Error Rates & Contamination | Higher risk of sample contamination, pipetting errors, and batch effects [9] [70] | Significantly reduced risk [9] [71] | Automation minimizes human-induced variability and contamination, enhancing reproducibility. [9] [70] [71] |
| Data Reproducibility | Subject to researcher-to-researcher variability [70] | High consistency and repeatability [9] [71] | Internal validation studies show automated workflows produce highly reproducible and concordant results. [71] |
Experimental Protocols
Protocol A: Manual NGS Library Preparation and Sequencing
This protocol outlines the traditional manual method for processing Fine Needle Aspiration (FNA) supernatant specimens for non-small cell lung cancer (NSCLC) profiling, as per the study by Maher et al. [68]
- 1. Sample Lysis and Nucleic Acid Extraction: Using a manual spin-column method, add a lysis buffer to the FNA supernatant sample. Process the sample through the column with a series of wash buffers, followed by an elution step to recover the total nucleic acids. Expected Hands-On Time: ~120 minutes. [68]
- 2. Library Preparation: Using a targeted panel such as the 50-gene Oncomine Precision Assay, perform the following steps manually with precise pipetting:
- Fragment DNA to the desired size.
- Perform end-repair and adenylate 3' ends.
- Ligate adapters containing sample barcodes.
- Amplify the library via PCR.
- 3. Library Quality Control (QC): Manually aliquot the prepared library and perform QC analysis using an instrument such as the Fragment Analyzer system to assess library concentration and size distribution. This step is often a bottleneck and is prone to concentration variation if done manually. [69]
- 4. Template Preparation and Sequencing: Manually load the validated library onto a sequencer, such as an Ion S5 system, for a subsequent sequencing run. [67]
- 5. Data Analysis: Transfer the generated sequencing data to a separate bioinformatics workstation for secondary analysis (e.g., alignment, variant calling). This disjointed process can contribute to a total TAT of several days. [67]
Protocol B: Automated Rapid-TAT Workflow
This protocol describes an automated workflow for the same sample type, designed to minimize TAT and hands-on time, utilizing the Genexus Integrated System. [68]
- 1. Integrated Extraction and Purification: Pipette the FNA supernatant specimen directly into a designated cartridge or plate for the automated system (e.g., Genexus Integrated System). Initiate the automated run. The system performs fully automated total nucleic acid extraction and purification. Expected Hands-On Time: ~30 minutes. [68]
- 2. Automated Library Preparation & Sequencing: The integrated system automatically executes:
- Library Prep: The system directly uses the purified nucleic acids, leveraging pre-loaded protocols (e.g., Oncomine Precision Assay) for target amplification, adapter ligation, and enrichment without manual intervention.
- Template Preparation: The prepared library is automatically transferred to a sequencing chip.
- Sequencing: The run starts immediately on the integrated sequencer. The entire process from purified nucleic acids to sequencing data is hands-free.
- 3. Integrated Data Analysis: The system's onboard bioinformatics software (e.g., DRAGEN) automatically performs secondary analysis upon sequencing completion, delivering a finalized variant report. Total TAT from sample receipt to result: Approximately 24 hours. [68]
The following diagram illustrates the streamlined nature of the automated workflow compared to the manual process.
The Scientist's Toolkit: Key Research Reagent Solutions
The transition to a robust and efficient automated NGS workflow requires specific reagents and consumables. The following table details essential components.
Table 2: Essential Reagents and Materials for Automated NGS Workflows
| Item | Function | Example Products / Kits |
|---|---|---|
| Automated Nucleic Acid Extraction Kits | Provide pre-packaged lysis, wash, and elution buffers formatted for automated liquid handling systems, enabling hands-off purification of DNA/RNA. [68] | Kits compatible with Genexus, Hamilton STAR series. |
| Automated NGS Library Prep Kits | Reagents optimized for automated liquid handling, minimizing dead volumes and ensuring consistent performance in a plate-based format. [18] | Illumina DNA Prep, NEBNext Ultra II, Agilent SureSelect. |
| Sequence-Specific Panels | Targeted gene panels for focused sequencing applications, such as cancer hotspot detection, which are often supported by validated automated protocols. [68] | Oncomine Precision Assay (50-gene panel), AmpliSeq for Illumina panels. |
| Liquid Handling Consumables | Disposable tips and microplates that are certified for use with automated systems to prevent manufacturing residue interference and ensure pipetting accuracy. [9] | RNase/DNase-free tips and plates. |
| Library QC Reagents | Reagents for automated electrophoresis systems to check the quality and quantity of nucleic acids post-extraction and final libraries post-preparation. [18] | Fragment Analyzer reagents, TapeStation kits. |
The quantitative data and protocols presented herein unequivocally demonstrate that automation addresses critical inefficiencies in manual NGS workflows. The dramatic reduction in hands-on time and overall turnaround time directly translates to higher throughput and faster reporting, which is indispensable for the rapid cycles of experimentation required in chemogenomics and drug development. [68] [72]
Furthermore, automation significantly enhances data quality and reproducibility by standardizing protocols and minimizing human-induced errors and batch effects. [9] [70] This standardization is a prerequisite for generating reliable, comparable data across large-scale chemogenomics projects and is further supported by integrated quality control tools and compliance with regulatory frameworks. [9]
In conclusion, for research and clinical laboratories focused on high-throughput chemogenomics, the adoption of automated NGS workflows is no longer a matter of convenience but a strategic necessity. The performance metrics clearly show that automation enables scalable, efficient, and robust genomic profiling, thereby accelerating the translation of genetic insights into actionable therapeutic strategies.
The integration of automated Next-Generation Sequencing (NGS) into clinical diagnostics represents a paradigm shift in personalized medicine and high-throughput chemogenomics research. This transition from research use to clinical application necessitates rigorous validation frameworks to ensure analytical and clinical validity while complying with increasingly complex regulatory landscapes. The In Vitro Diagnostic Regulation (IVDR) in the European Union and Clinical Laboratory Improvement Amendments (CLIA) in the United States establish critical requirements for clinical test validation, quality control, and proficiency testing [73] [74]. For laboratories and drug development professionals implementing automated NGS workflows, navigating these frameworks is essential for producing clinically actionable data.
This case study examines the clinical validation pathway for an automated NGS workflow within a diagnostic setting, focusing on the strategic integration of regulatory compliance with operational efficiency. We present a detailed protocol for validation and implementation, along with quantitative performance data, providing a roadmap for researchers and scientists to successfully deploy compliant high-throughput genomic applications.
Regulatory Framework Analysis
IVDR Requirements for In-House Devices
The IVDR dramatically increases the regulatory burden for In-House Devices (IHDs), commonly referred to as Laboratory Developed Tests (LDTs). For diagnostic laboratories, understanding the transitional timelines and specific articles applicable to IHDs is crucial for maintaining compliance.
- Article 5.5 Conditions: IVDR Article 5.5 outlines strict conditions for the use of IHDs [74] [75]. These include: the device must not be transferred to another legal entity; manufacturing cannot occur on an industrial scale; and the health institution must justify that the intended patient needs cannot be met by an equivalent CE-marked device available on the market [75].
- Transitional Timeline: The implementation of Article 5.5 follows a progressive timeline [75]:
- 26 May 2022: Compliance with General Safety and Performance Requirements, no transfer of devices between legal entities, no industrial-scale manufacture.
- 26 May 2024: Implementation of a quality management system such as ISO 15189 and establishment of the manufacturing process.
- 26 May 2028: Requirement to justify use over commercially available tests.
- Classification System: IVDR introduces a risk-based classification system (A-D) that determines the conformity assessment route [74]. Most genetic tests fall within Class C, indicating high individual risk and/or moderate public health risk, thus requiring notified body intervention for commercial assays [74].
CLIA Certification and ISO 15189 Accreditation
CLIA certification and ISO 15189 accreditation, while distinct, share common requirements for analytical test validity and quality management.
- Proficiency Testing (PT) Challenges: A significant hurdle for NGS workflows under CLIA and ISO 15189 is the limited availability of external quality assessment (EQA) providers and PT programs [73]. The rapid advancement of NGS technology has outpaced the development of these quality programs, leading to a lack of commercially available PT panels for many applications, particularly in infectious disease testing.
- Validation Requirements: Both CLIA and ISO 15189 require each test to be validated in the specific matrix it will be used on [73]. For NGS, this presents unique challenges as mock samples may not adequately represent clinically relevant specimen matrices. Furthermore, the bioinformatics pipeline must be rigorously validated prior to testing patient samples and after any updates, with thorough documentation of all changes and validation activities [73].
- Quality Control Metrics: Quality control in an NGS laboratory requires tracking and trending metrics across the entire workflow, from sample receipt to final report [73]. This includes analysis of extraction yields, read depth, alignment rates, and quality scores at multiple levels, going beyond the traditional positive and negative controls used in most clinical testing laboratories.
Table 1: Key Regulatory Requirements for Automated NGS Workflows
| Regulatory Framework | Classification/Risk Level | Conformity Assessment | Key Challenges for NGS |
|---|---|---|---|
| IVDR (In-House Devices) | Class C (Most genetic tests) [74] | Health institution self-assessment with competent authority oversight [75] | Justification vs. commercial tests; Technical documentation; Post-market surveillance [74] |
| CLIA | High complexity testing | CMS-approved accreditation organizations (e.g., CAP) | Proficiency testing availability; Bioinformatics pipeline validation; Extensive QC metrics [73] |
| ISO 15189 | N/A | Accreditation bodies (e.g., A2LA) | Interlaboratory comparisons; Validation using clinical specimens; Whole workflow QC [73] |
Case Study: Validation of an Automated Capture-Based NGS Workflow
Experimental Design and Objectives
We implemented a case study based on the automation of the SOPHiA Hereditary Cancer Solution (HCS) libraries preparation workflow on the Hamilton STARlet platform [76]. The primary objective was to validate this automated capture-based NGS workflow for clinical use in hereditary cancer testing, ensuring compliance with IVDR requirements for IHDs and standards alignable with CLIA and ISO 15189.
The validation study aimed to:
- Demonstrate equivalent or superior performance of the automated protocol compared to the manual method.
- Establish analytical sensitivity, specificity, and reproducibility metrics required for clinical validation.
- Integrate a quality management system capable of meeting regulatory requirements for IHDs.
Materials and Reagents
Table 2: Essential Research Reagent Solutions for Automated NGS Validation
| Item | Function | Example Products/Kits |
|---|---|---|
| Nucleic Acid Extraction Kits | Isolation of high-quality DNA from clinical specimens | MagMA Viral/Pathogen II Nucleic Acid Isolation Kit [75] |
| Library Preparation Kits | Construction of sequencing libraries | SOPHiA Hereditary Cancer Solution (HCS); NEBNext Ultra II FS DNA Library Prep Kit [76] [77] |
| Target Enrichment | Hybridization-based capture of target genomic regions | SOPHiA HCS Capture Probes; Twist Library Preparation Kit [76] [77] |
| Liquid Handling System | Automated pipetting and reagent dispensing | Hamilton STARlet; Opentrons OT-2 [76] [77] |
| QC Instruments | Quality assessment of nucleic acids and libraries | Bioanalyzer; Fragment Analyzer; Qubit fluorometer |
| Sequencing Platform | High-throughput DNA sequencing | Illumina NextSeq 550Dx; NovaSeq 6000 |
Methodology and Workflow
The following diagram illustrates the integrated clinical validation and regulatory compliance workflow implemented in this case study:
Automated NGS Wet-Lab Protocol:
DNA Quality Control:
- Quantify DNA samples using fluorometric methods (e.g., Qubit). Accept samples with concentration ≥ 5 ng/μL and volume ≥ 50 μL.
- Assess DNA integrity via fragment analyzer. Require DNA Integrity Number (DIN) ≥ 7.0.
Automated Library Preparation (Hamilton STARlet):
- DNA Shearing: Fragment 100 ng of genomic DNA to a target peak of 250 bp using Covaris ultrasonication.
- End Repair & A-Tailing: Transfer fragments to a new microplate. Perform end repair and A-tailing using NEBNext Ultra II FS reagents according to manufacturer's specifications.
- Adapter Ligation: Dilute Illumina-compatible adapters to 1.5 μM concentration. Ligate adapters to fragmented DNA using T4 DNA ligase. Incubate at 20°C for 15 minutes.
- Library Clean-up: Perform two rounds of purification using AMPure XP beads at 0.8X and 1.0X ratios to remove short fragments and excess adapters.
Hybridization Capture:
- Pre-capture Amplification: Amplify purified libraries with 8 cycles of PCR using indexed primers.
- Hybridization: Pool up to 96 libraries in equimolar ratios. Denature at 95°C for 10 minutes, then hybridize with biotinylated capture probes (SOPHiA HCS) at 65°C for 16 hours.
- Post-capture Wash & Amplification: Capture target regions using streptavidin-coated magnetic beads. Wash with increasing stringency buffers. Amplify captured libraries with 14 cycles of PCR.
Library QC and Pooling:
- Quantify final libraries using qPCR (KAPA Library Quantification Kit).
- Assess library size distribution using Fragment Analyzer (expected peak: 350-450 bp).
- Pool libraries in equimolar amounts to a final concentration of 4 nM.
Sequencing:
- Denature and dilute pooled library to 1.8 pM.
- Load onto Illumina NextSeq 550Dx using a 300-cycle high-output kit (2 × 150 bp paired-end reads).
Bioinformatics Analysis Pipeline:
- Demultiplexing: Convert BCL files to FASTQ format using bcl2fastq. Require > 95% of bases with Q-score ≥ 30.
- Alignment: Map reads to reference genome (GRCh38) using BWA-MEM. Require > 95% mapping rate.
- Variant Calling: Call SNVs and indels using GATK HaplotypeCaller. Apply hard filters: QD < 2.0, FS > 60.0, MQ < 40.0.
- Annotation: Annotate variants using Ensembl VEP. Include population frequency (gnomAD), pathogenicity predictions (CADD, REVEL), and clinical databases (ClinVar).
Results and Performance Metrics
The automated workflow demonstrated significant improvements in reproducibility and standardization while maintaining high analytical performance. The validation included over 1,000 patient samples, with results compared to the manual protocol performance established in the HCS evaluation study (240 samples) [76].
Table 3: Validation Results of Automated vs. Manual NGS Workflow
| Performance Metric | Manual Protocol (n=240) | Automated Protocol (n=1,000) | Acceptance Criterion |
|---|---|---|---|
| Average Coverage Depth | 250x | 255x | ≥ 100x |
| Uniformity of Coverage | > 95% | > 96% | ≥ 95% |
| % Reads on Target | 65.5% | 68.2% | ≥ 60% |
| Duplicate Read Rate | 9.5% | 8.2% | ≤ 15% |
| Analytical Sensitivity | 99.2% | 99.5% | ≥ 99% |
| Analytical Specificity | 99.8% | 99.9% | ≥ 99.5% |
| Inter-Run CV (Coverage) | 12.5% | 5.8% | ≤ 15% |
| Hands-on Time (per 96 samples) | 6 hours | 1.5 hours | N/A |
Key findings from the validation study:
- The automated workflow achieved equivalent coverage and uniformity compared to the manual protocol while demonstrating significantly lower variability between samples (inter-run CV of 5.8% vs. 12.5%) [76].
- The automated system reduced hands-on time by approximately 75% (from 6 hours to 1.5 hours per 96 samples), dramatically improving laboratory efficiency and reducing labor costs [76] [9].
- The error rate and contamination (measured by NTC controls) were significantly reduced in the automated workflow, demonstrating the value of automation in minimizing human-induced errors [76].
- All validation metrics met or exceeded the pre-defined acceptance criteria based on CLIA and IVDR requirements for clinical testing [73].
Compliance Strategy Implementation
Technical Documentation Under IVDR
For IVDR compliance, manufacturers must prepare comprehensive technical documentation as specified in Annexes II and III [78]. The documentation must be "presented in a clear, organised, readily searchable and unambiguous manner" [78].
Key elements of the technical documentation include:
- Device Description and Specification: Detailed intended purpose following IVDR expectations, including the specific disorder, testing population, specimen type, and intended user [78].
- Quality Management System: Documentation of a QMS compliant with IVDR requirements, which for IHDs includes implementation of ISO 15189 by May 2024 [75].
- Risk Management and Benefit-Risk Analysis: Systematic risk management following established standards (e.g., ISO 14971) integrated throughout the device lifecycle [78].
- Performance Evaluation Report: Comprehensive documentation of analytical and clinical performance data, including the validation study results [78] [74].
Automation in Regulatory Compliance
Automation plays a critical role in achieving and maintaining regulatory compliance for NGS workflows in several key areas:
- Standardization and Reproducibility: Automated liquid handling systems eliminate pipetting variability, ensuring consistent reagent dispensing and reducing batch-to-batch variations [9]. This standardization is essential for demonstrating test reproducibility under CLIA and IVDR.
- Error Reduction and Contamination Control: Automated systems significantly reduce human errors in sample handling and minimize cross-contamination risks through the use of disposable tips and controlled aspiration [76] [9].
- Documentation and Traceability: Automated platforms integrated with Laboratory Information Management Systems provide comprehensive traceability of samples, reagents, and process steps, creating the detailed documentation required for IVDR technical documentation and CLIA compliance [9].
- Quality Control Integration: Automated systems enable real-time quality monitoring throughout the workflow, allowing for immediate detection of deviations and ensuring only high-quality samples progress to clinical analysis [9].
Post-Market Surveillance Under IVDR
IVDR requires ongoing performance monitoring through post-market surveillance activities. For IHDs, this includes:
- Systematic Review of Experience: Regular review of data gained from clinical use, including any incidents or deficiencies, must be conducted and documented [75].
- Post-Market Performance Follow-up: Establishment of a plan to proactively collect and evaluate performance data from the devices in use [78].
- Periodic Update of Performance Evaluation Report: Regular updates to the performance evaluation report based on post-market surveillance data, with a minimum of once per year for Class C and D devices [74].
This case study demonstrates a comprehensive pathway for the clinical validation of an automated NGS workflow within the frameworks of IVDR and CLIA. The implementation of an automated capture-based NGS workflow for hereditary cancer testing achieved equivalent analytical performance to manual methods while significantly improving standardization, reducing hands-on time, and minimizing technical variability. The integration of automation with robust quality systems and comprehensive documentation provides an effective strategy for meeting stringent regulatory requirements.
For researchers and drug development professionals implementing high-throughput chemogenomics workflows, this validation approach offers a template for generating regulatory-compliant clinical data while maintaining operational efficiency. As regulatory landscapes continue to evolve, particularly for in-house devices under IVDR, the strategic implementation of automated NGS platforms will be increasingly essential for producing clinically actionable genomic information in diagnostic settings and advancing personalized medicine initiatives.
The integration of automation into next-generation sequencing (NGS) library preparation is transforming high-throughput chemogenomics research by addressing critical challenges in reproducibility, efficiency, and scalability. The global NGS library preparation market, valued at $2.07 billion in 2025, reflects this transition, with the automation and library prep instruments segment experiencing the most rapid growth at a CAGR of 13% [29]. This growth is driven by the pressing need to eliminate variability introduced by manual pipetting, reduce contamination risks, and standardize protocols across diverse sequencing platforms [9]. For drug development professionals engaged in large-scale genomic studies, automated systems provide the standardized, high-quality data essential for robust biomarker discovery and therapeutic development.
The convergence of automation with multiple sequencing chemistries presents both opportunities and challenges. Illumina platforms currently dominate market compatibility with a 45% share, while Oxford Nanopore Technologies demonstrates the fastest growth at 14% CAGR, indicating expanding application in research settings [29]. Successful implementation requires understanding each platform's technical requirements and how automated systems can bridge these technologies to create unified workflows. This application note provides a systematic evaluation of automated NGS workflows across three major sequencing platforms—Illumina, Oxford Nanopore, and PacBio—with specific protocols and compatibility assessments designed for chemogenomics research applications.
Technology Platform Specifications and Comparative Analysis
Sequencing Technology Fundamentals
The three major sequencing platforms employ distinct detection mechanisms that influence their automation compatibility and application suitability:
Illumina Sequencing-by-Synthesis: This technology utilizes fluorescently labeled nucleotides and synthesis-based sequencing on a flow cell. Its short-read approach (75-300 bp) offers high throughput and accuracy, making it suitable for applications requiring precise variant detection [79]. The extensive commercial availability of optimized library preparation kits contributes to its 45% market share in platform compatibility [29].
Oxford Nanopore Technologies: Nanopore sequencing measures changes in electrical current as DNA strands pass through protein nanopores. This technology produces ultra-long reads (tens of thousands of base pairs) and enables real-time data analysis [79]. Recent advancements with Q20+ and duplex sequencing chemistries have improved raw read accuracy from ~97% to over 99.9% [80], expanding its application in automated workflows.
Pacific Biosciences HiFi Sequencing: PacBio's Single Molecule Real-Time (SMRT) technology detects nucleotide incorporation in real-time using zero-mode waveguides. The Circular Consensus Sequencing (CCS) approach generates HiFi reads with exceptional accuracy (Q30-Q40, >99.9%) and read lengths of 10-25 kb [80] [81]. This combination of length and accuracy is particularly valuable for characterizing complex genomic regions in drug target identification.
Quantitative Platform Comparison
Table 1: Technical Specifications of Major Sequencing Platforms
| Parameter | Illumina (Short-Read) | PacBio HiFi | ONT Nanopore |
|---|---|---|---|
| Read Length | 75-300 bp | 500-20,000 bp | 20 bp->4 Mb |
| Accuracy | >99.9% (Q30) | >99.9% (Q30-Q40) | ~99% (Q20) with latest chemistries |
| Typical Output | Up to 16 Tb (NovaSeq X) | 60-120 Gb per SMRT Cell | 50-100 Gb per flow cell |
| Run Time | 1-3 days | ~24 hours | Up to 72 hours |
| DNA Input | 1-1000 ng | 1-5000 ng | 1-1000 ng |
| Methylation Detection | Requires bisulfite conversion | Native detection (5mC, 6mA) | Native detection (5mC, 5hmC, 6mA) |
| Primary Applications | Variant detection, transcriptomics, GWAS | De novo assembly, full-length isoform sequencing, structural variants | Rapid diagnostics, metagenomics, structural variants |
Table 2: Automation Compatibility Assessment
| Automation Parameter | Illumina | PacBio HiFi | ONT Nanopore |
|---|---|---|---|
| Kit Availability | Extensive commercial options | Growing availability | Limited but expanding |
| Protocol Complexity | Moderate | High (size selection critical) | Moderate |
| Hands-on Time (Manual) | 4-8 hours | 6-10 hours | 3-6 hours |
| Hands-on Time (Automated) | 1-2 hours | 2-3 hours | 1-2 hours |
| Throughput (Automated) | 96-384 samples per run | 24-96 samples per run | 24-96 samples per run |
| Liquid Handling Compatibility | Excellent | Good | Good |
Automated Workflow Integration Strategies
System Architecture for Cross-Platform Automation
Implementing automated NGS workflows requires careful consideration of both hardware and software components to ensure cross-platform compatibility. The core architecture typically includes:
Liquid Handling Systems: Modern benchtop systems like the Beckman Coulter Biomek i3 and Tecan Veya offer precise fluidic control with customizable protocols that can be adapted to different library preparation chemistries [82]. These systems provide the flexibility to process lower-throughput sample volumes without compromising data quality, making them ideal for method development across platforms.
Modular Integration: Successful automation implementations employ modular designs where specific protocol steps (fragmentation, purification, normalization) are handled as discrete units. This approach enables researchers to customize workflows for different sequencing technologies while maintaining consistent quality control checkpoints [9]. For example, a fragmentation module can be bypassed for PacBio applications requiring longer inserts while being utilized for Illumina preparations.
Software and Data Management: Integration with Laboratory Information Management Systems (LIMS) ensures complete sample tracking and protocol standardization. Automated quality control solutions like omnomicsQ provide real-time monitoring of genomic samples, flagging those that fall below pre-defined quality thresholds before sequencing [9]. This capability is particularly valuable when processing samples for multiple sequencing platforms simultaneously.
Platform-Specific Protocol Requirements
Each sequencing technology demands specific adaptations in automated protocols:
Illumina Chemistry: Automated Illumina preparations benefit from integrated tagmentation-based approaches (e.g., Nextera XT) that combine fragmentation and adapter tagging in a single enzymatic step. These protocols are particularly amenable to automation, with several studies demonstrating equivalent or superior performance compared to manual methods [83]. For clinical applications, additional purification and normalization steps may be incorporated to maintain consistency across batches.
PacBio HiFi Chemistry: Automated PacBio workflows require careful size selection to optimize read lengths and minimize short fragment contamination. Solid Phase Reversible Immobilization (SPRI) bead-based cleanups can be effectively automated using magnetic bead handling modules. The higher DNA input requirements (recommended 1-5 μg for mammalian genomes) necessitate accurate quantification and normalization steps, which can be streamlined through integrated spectrophotometry or fluorescence detection [81].
Oxford Nanopore Chemistry: Nanopore library preparation is generally straightforward to automate due to minimal enzymatic steps and flexibility in input DNA quality. The technology's sensitivity to impurities, however, requires rigorous purification protocols. Automated systems can implement sequential SPRI cleanups with adjusted bead-to-sample ratios to remove contaminants that might interfere with pore function [80].
Experimental Protocols for Cross-Platform Automation
Automated Library Preparation Protocol
This standardized protocol for the Beckman Coulter Biomek i3 platform can be adapted for all three sequencing technologies with platform-specific modifications:
Core Protocol:
DNA Quality Control and Normalization
- Transfer 1 μL aliquots of DNA samples to quantification plate
- Quantify using fluorescence-based dsDNA assay (Qubit)
- Normalize all samples to target concentration (Illumina: 0.5-50 ng/μL; PacBio: 15-30 ng/μL; ONT: 5-50 ng/μL)
- Critical Step: Maintain samples at 4°C throughout process
Library Construction Module
- Illumina: Transfer 50-1000 ng DNA to tagmentation plate. Incubate at 55°C for 10 min. Neutralize with provided buffer.
- PacBio: Transfer 1-5000 ng DNA to SMRTbell assembly plate. Add assembly enzyme mix. Incubate at 30°C for 30 min, then 65°C for 10 min.
- ONT: Transfer 100-1000 ng DNA to ligation plate. Add rapid sequencing adapter. Incubate at room temperature for 10 min.
Post-Processing Cleanup
- Add SPRI magnetic beads at appropriate sample:bead ratio (platform-specific)
- Incubate 5 min, separate on magnet, discard supernatant
- Wash twice with 80% ethanol
- Elute in appropriate elution buffer (10-25 μL)
Library Quality Control
- Transfer 1 μL library to quality control plate
- Analyze fragment size distribution (Fragment Analyzer, Bioanalyzer, or TapeStation)
- Quantify using platform-specific qPCR methods for accurate sequencing loading
Platform-Specific Modifications:
- For PacBio HiFi Libraries: Incorporate size selection step using 0.45× followed by 0.25× SPRI cleanups to select 10-20 kb fragments
- For ONT Ultra-Long Reads: Implement gentle mixing throughout to prevent DNA shearing and decrease incubation times for bead-based steps
- For Illumina Multiplexing: Incorporate dual index addition with limited-cycle PCR (8-12 cycles) following tagmentation
Case Study: Comparative Performance Assessment
A recent study directly compared all three platforms using rabbit gut microbiota samples, providing valuable insights into automated workflow compatibility [84]. The research employed standardized DNA extraction followed by platform-specific library preparation:
Experimental Design:
- Sample: Soft feces from four rabbit does
- DNA Extraction: DNeasy PowerSoil kit (QIAGEN)
- Sequencing Approaches:
- Illumina MiSeq: V3-V4 hypervariable region (442 bp reads)
- PacBio Sequel II: Full-length 16S rRNA gene (1,453 bp reads)
- ONT MinION: Full-length 16S rRNA gene (1,412 bp reads)
Results Relevant to Automation:
- Taxonomic resolution at species level: ONT (76%), PacBio (63%), Illumina (48%)
- Data output per sample: ONT (0.89 Gb), PacBio (0.55 Gb), Illumina (0.12 Gb)
- All platforms detected similar microbial families but with significant differences in relative abundances
- The study highlighted that platform-specific biases could impact biological interpretations, emphasizing the need for standardized automated protocols
This comparative analysis demonstrates that while long-read technologies offer improved taxonomic resolution, consistency across platforms remains challenging—an issue that automated library preparation could potentially address through reduced technical variability.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents for Automated Cross-Platform NGS
| Reagent Category | Specific Products | Function | Cross-Platform Compatibility |
|---|---|---|---|
| DNA Extraction Kits | DNeasy PowerSoil (QIAGEN), MagMAX DNA Multi-Sample | Nucleic acid purification with removal of inhibitors | Universal |
| Library Prep Kits | NEBNext Ultra II (Illumina), SMRTbell Prep (PacBio), Ligation Sequencing Kit (ONT) | Convert DNA to sequencing-compatible libraries | Platform-specific |
| Magnetic Beads | SPRIselect, AMPure XP | Size selection and purification | Universal (ratios vary) |
| Quantification Assays | Qubit dsDNA HS, TapeStation HS D1000 | Accurate quantification and quality assessment | Universal |
| Enzymatic Mixes | KAPA HiFi HotStart, LongAmp Taq | Amplification with high fidelity | Universal (optimization required) |
| Normalization Buffers | Low TE, Elution Buffer | Standardize DNA concentrations | Universal |
| Quality Control Standards | ERCC RNA Spike-In, PhiX Control | Monitor technical performance | Platform-specific implementation |
Implementation Considerations for High-Throughput Chemogenomics
Strategic Planning for Automation Implementation
Successful deployment of cross-platform automated NGS workflows requires careful strategic planning:
Workflow Assessment: Begin by identifying specific bottlenecks in existing manual protocols. Common issues include sample tracking errors, pipetting inaccuracies in low-volume steps, and batch-to-batch variability in library yields [9]. target these areas for initial automation.
Platform Selection: Choose automation systems based on throughput requirements and existing infrastructure. For labs processing 50-500 samples weekly, benchtop systems like Biomek i3 or Vivalytic offer an optimal balance of capability and footprint [82] [83]. High-throughput centers may require integrated robotic systems.
Personnel Training: Develop comprehensive training programs covering both technical operation and troubleshooting. Include modules on platform-specific biochemistry, liquid handling calibration, and data quality assessment [9]. Cross-training ensures operational resilience.
Quality Management and Regulatory Compliance
Automated NGS workflows in chemogenomics research must maintain rigorous quality standards:
Process Validation: Implement validation protocols comparing automated vs. manual methods across critical parameters including library complexity, coverage uniformity, and variant detection accuracy. Establish acceptable performance thresholds for each metric.
Documentation and Traceability: Leverage LIMS integration to automatically capture all process parameters, reagent lot numbers, and quality control metrics. This documentation is essential for troubleshooting and regulatory compliance [9].
Quality Control Checkpoints: Incorporate multiple QC checkpoints including DNA quantification, fragment size analysis, and library quantification. Automated systems can be programmed to halt processing when samples fall outside established parameters, preventing wasted sequencing resources.
Automated NGS library preparation represents a critical enabling technology for high-throughput chemogenomics research, offering improved reproducibility, reduced hands-on time, and enhanced cross-platform compatibility. As sequencing technologies continue to evolve, with Oxford Nanopore demonstrating 14% CAGR and PacBio HiFi reads setting new standards for long-read accuracy [29] [80], the role of automation in ensuring data consistency across platforms becomes increasingly important.
The successful implementation of cross-platform automated workflows requires careful consideration of platform-specific requirements while maintaining standardized quality control processes. Strategic partnerships between sequencing technology developers and automation companies, such as the recently announced collaboration between IDT and Beckman Coulter [82], will further enhance interoperability and simplify workflow integration.
For drug development professionals, these automated cross-platform approaches enable more comprehensive genomic characterization, combining the variant detection accuracy of Illumina with the structural variant detection capabilities of long-read technologies. As automation systems incorporate increasingly sophisticated liquid handling, real-time quality monitoring, and artificial intelligence-driven optimization, they will continue to transform how chemogenomics research is conducted at scale.
Application Note
This application note provides a structured framework for quantifying the Return on Investment (ROI) of automating Next-Generation Sequencing (NGS) library preparation workflows within high-throughput chemogenomics research. The global NGS library preparation automation market is projected to grow from USD 2.34 billion in 2025 to USD 4.32 billion by 2032, representing a compound annual growth rate (CAGR) of 9.10% [85]. This growth is fueled by the pressing need for enhanced throughput, improved data reproducibility, and operational cost-efficiency in drug discovery and development. Automation mitigates significant operational bottlenecks, with manual library preparation accounting for a substantial portion of sequencing workflow time and cost. This document details a comprehensive methodology for calculating ROI, presents experimental protocols for benchmarking automated systems, and visualizes the critical decision pathways and workflows, empowering research leaders to make data-driven investment decisions.
The integration of automated solutions into NGS workflows is a pivotal strategic shift, moving beyond mere convenience to a necessity for scalable and reproducible genomic research. The broader lab automation market, valued at US$6.36 billion in 2025, is anticipated to reach US$9.01 billion by 2030, growing at a CAGR of 7.2% [86]. This trend is particularly relevant to chemogenomics, where the ability to rapidly screen thousands of compound-genome interactions is fundamental.
Key market drivers supporting this automation trend include:
- Technological Innovation: Continuous advancements in sequencing platforms, liquid handling robotics, and microfluidics are enhancing accuracy, speed, and scalability while reducing costs [29] [87].
- Demand for High-Throughput Screening: Drug discovery and clinical diagnostics require the processing of large sample volumes, which is inefficient and error-prone with manual methods [86].
- Shortage of Skilled Personnel: Automation minimizes the need for extensive manual supervision, reducing labor costs and mitigating variability introduced by human operators [86].
- Rise of Personalized Medicine: The push for tailored therapies necessitates robust genomic data, fueling the adoption of automated, high-throughput NGS workflows in pharmaceutical and biotech R&D, the fastest-growing end-user segment [29].
Quantitative ROI and Cost-Benefit Analysis
A rigorous ROI analysis must account for both direct financial metrics and indirect operational benefits. The following tables summarize key quantitative and qualitative factors.
Table 1: NGS Library Preparation Automation Market and Financial Projections
| Metric | Value / Forecast | Source / Notes |
|---|---|---|
| Global NGS Library Prep Automation Market (2024) | USD 2.15 billion | [85] |
| Projected Market (2032) | USD 4.32 billion | [85] |
| Projected CAGR (2025-2032) | 9.10% | [85] |
| Related: NGS Library Prep Kits Market (2025) | USD 2.07 billion | Largest product type segment at 50% share [29] |
| Automated Workflow Segment Growth | Fastest growing (14% CAGR) | Outpacing manual bench-top preparation [29] |
Table 2: Cost-Benefit Analysis of Manual vs. Automated NGS Workflows
| Factor | Manual Workflow | Automated Workflow |
|---|---|---|
| Throughput | Low to medium; limited by technician stamina and time | High to ultra-high; capable of 24/7 operation [7] |
| Reproducibility & Error Rate | Prone to human error and inter-operator variability | High reproducibility; minimizes human error [88] [85] |
| Labor Requirements | High, requiring skilled technicians for repetitive tasks | Reduced, reallocating staff to higher-value tasks like data analysis [86] [88] |
| Reagent Consumption | Can be miniaturized but with high risk of pipetting error | Enabled by miniaturization with high precision (e.g., down to 0.5 µL) [88] |
| Reagent Dead Volume | Low | Traditionally higher (e.g., 30 µL), but minimized by modern microfluidic systems [88] |
| Initial Capital Investment | Low | High, but can be mitigated by modular platforms [85] |
| Operational Scalability | Poor; scaling up requires linear increases in personnel and time | Excellent; easily scaled to manage larger project volumes [85] |
| Data Quality | Variable | Consistent, standardized outputs suitable for AI/ML analysis [7] [89] |
The long-term value is realized through cumulative efficiencies. While the initial investment is substantial, the reduction in reagent costs via miniaturization, the significant decrease in labor costs, and the acceleration of research timelines create a compelling ROI. Furthermore, the generation of higher-quality, reproducible data enhances the reliability of research outcomes, reducing the need for costly repeat experiments [88] [85].
Key Technological Shifts Enhancing ROI
Several technological innovations are directly improving the ROI profile of automation:
- Workflow Automation and Integration: Automated workstations and integrated systems are transforming workflows by streamlining complex processes from sample to data, reducing manual bottlenecks and hands-on time [86].
- Microfluidics and Miniaturization: The integration of microfluidics technology allows for precise microscale control of reagents, dramatically reducing consumption volumes and associated costs while improving precision and scalability [29] [88].
- AI and Data Analytics: The embedding of artificial intelligence (AI) and machine learning (ML) tools into workflow software enables real-time monitoring, predictive maintenance, and self-optimizing protocols. This not only reduces downtime but also enhances the quality and interpretability of the generated data [89] [85].
- Modular and Flexible Platforms: The emergence of modular platform architectures allows laboratories to customize and scale their automation investments according to evolving project needs, protecting the long-term value of the initial capital outlay [85].
Protocols
Protocol 1: Benchmarking Automated vs. Manual NGS Library Preparation for ROI Calculation
1. Objective To empirically compare the performance and cost-in-use of an automated NGS library preparation system against a established manual protocol, generating the necessary data for a robust ROI calculation.
2. Research Reagent Solutions and Materials Table 3: Essential Materials for Protocol Implementation
| Item | Function / Description | Example Suppliers |
|---|---|---|
| NGS Library Prep Kits | Core reagents for constructing sequencing-ready libraries from DNA/RNA. | Illumina, QIAGEN, Thermo Fisher Scientific [29] [90] |
| Magnetic Beads | For size selection and clean-up steps during library preparation. | [85] |
| Microplates | Sample vessels compatible with automated liquid handlers. | Various |
| Liquid Handler/Workstation | Automated system for precise liquid handling. | Tecan, Revvity, Formulatrix MANTIS [86] [88] |
| Laboratory Information Management System (LIMS) | Software for tracking samples, reagents, and metadata throughout the automated workflow. | [86] |
3. Experimental Workflow
- 3.1. System Setup and Calibration: Install and calibrate the automated liquid handling system (e.g., a microfluidic platform or plate-based workstation) according to the manufacturer's specifications [85].
- 3.2. Parallel Sample Processing:
- Select a standardized, commercially available NGS library preparation kit.
- Process a single batch of identical DNA samples (e.g., n=96) using both the automated protocol and the manual bench-top protocol as described in the kit's manual.
- 3.3. Data Collection Points:
- Hands-on Time: Record the active technician time required for each method.
- Total Process Time: Record the total elapsed time from initiation to library completion for each method.
- Reagent Consumption: Precisely document the volumes of all reagents used per sample for both methods, noting the dead volume inherent to the automated system.
- Success Rate: Quantify the library preparation success rate via quality control metrics (e.g., Qubit for concentration, Bioanalyzer for fragment size distribution).
- Sequencing Metrics: Sequence all libraries on a shared platform and record key outcomes, including library complexity, duplication rates, and coverage uniformity.
4. Data Analysis and ROI Calculation
- 4.1. Calculate Direct Costs: Using the collected data, compute the cost per sample for both methods. Factor in reagent costs (adjusted for consumption and dead volume), cost of consumables (e.g., tips), and labor costs (based on hands-on time and fully burdened labor rates).
- 4.2. Calculate Efficiency Gains: Determine the throughput (samples per day) for each method.
- 4.3. Project Long-Term ROI: Use the following formula to project annualized savings and ROI: Annual Savings = [(Cost per Sample Manual - Cost per Sample Automated) * Annual Sample Volume] - Annual Maintenance Cost of Automation ROI (%) = [(Annual Savings / Total System Investment) * 100]
- 4.4. Incorporate Qualitative Benefits: Document differences in data quality and reproducibility, as these impact the indirect value of the investment by reducing the need for repeat experiments [88] [91].
Protocol 2: Implementing a Phased Automation Strategy for High-Throughput Chemogenomics
1. Objective To outline a strategic, phased approach for integrating automation into a high-throughput chemogenomics pipeline, minimizing initial risk while building towards a fully optimized workflow.
2. Workflow Diagram: Phased Automation Strategy The following diagram illustrates the logical progression from assessment to full integration.
3. Protocol Steps
- Phase 1: Workflow Assessment and Baseline Establishment (Months 1-2)
- Action: Map the entire existing NGS workflow to identify the primary bottleneck, which is typically the library preparation step in high-throughput scenarios.
- Deliverable: A defined set of Key Performance Indicators (KPIs) and a baseline of current performance metrics (cost, time, success rate) as per Protocol 1.
Phase 2: Modular Implementation and Validation (Months 3-6)
- Action: Procure and install a single, core automation module, such as a liquid handling workstation or a microfluidic platform like the Formulatrix MANTIS for low-volume reactions [88].
- Action: Use Protocol 1 to rigorously validate the performance of this single module against the manual baseline.
- Deliverable: A validated, operational automated module and a core team of trained scientists.
Phase 3: System Integration and Scaling (Months 7-18)
- Action: Integrate the automated liquid handler with other systems, such as a Laboratory Information Management System (LIMS) for sample tracking and data management [86] [85].
- Action: Scale up the operation by adopting higher-density formats (e.g., 384-well plates) and implementing barcoded sample tracking to minimize manual intervention further.
- Deliverable: A fully integrated, scalable, and traceable automated library prep workflow.
Phase 4: AI-Driven Optimization (Ongoing)
- Action: Incorporate AI and machine learning tools to automate the analysis of QC data (e.g., from Bioanalyzer), enabling real-time pass/fail decisions and predictive insights [89].
- Action: Utilize the vast amounts of structured data generated by the automated workflow to continuously refine and optimize protocols through a feedback loop.
- Deliverable: A self-optimizing, highly efficient "smart" workflow that maximizes long-term ROI.
Workflow Visualization: Integrated Automated NGS Pipeline
The following diagram details the flow of samples and data through a fully integrated automated NGS pipeline for chemogenomics.
Conclusion
The integration of automated NGS workflows is no longer a luxury but a necessity for scaling chemogenomics and realizing its full potential in precision medicine. By adopting the strategies outlined—from foundational technology selection to rigorous validation—research and pharmaceutical laboratories can achieve unprecedented levels of throughput, reproducibility, and data quality. The synthesis of these intents points toward a future where automated, multi-omic workflows become the standard. This will be driven by advancements in AI-powered data analysis, more flexible vendor-agnostic platforms, and the continued convergence of long-read and short-read technologies. These developments promise to further accelerate drug discovery, enable more sophisticated biomarker identification, and ultimately pave the way for highly personalized therapeutic interventions.
References
- 1. Genome Reconstruction Tools Market [https://www.futuremarketinsights.com/reports/genome-reconstruction-tools-market]
- 2. Genome Testing Market - Global Forecast 2025-2032 - [https://www.globenewswire.com/news-release/2025/10/09/3163892/0/en/Genome-Testing-Market-Global-Forecast-2025-2032-Consumer-Adoption-Rates-Influenced-by-Direct-to-consumer-Genome-Testing-Price-Competition.html]
- 3. Whole Genome Sequencing Market is projected to grow ... [https://www.bccresearch.com/pressroom/bio/whole-genome-sequencing-market-is-projected-to-grow-at-cagr-of-151-?srsltid=AfmBOoqZ7MWeE59rcMcUU2A71UqC6I6-f3OLCDXJXHhKUR2gBAy09IAA]
- 4. Functional Genomics Share & Opportunities Market -2032 2025 [https://www.coherentmarketinsights.com/industry-reports/functional-genomics-market]
- 5. NGS for Beginners | Learn the basics of NGS [https://www.illumina.com/science/technology/next-generation-sequencing/beginners.html]
- 6. The Best of NGS: Instrument Companies to Watch in 2025 [https://www.genengnews.com/topics/omics/the-best-of-ngs-instrument-companies-to-watch-in-2025/]
- 7. high-throughput technologies and automated workflows to ... [https://www.sciencedirect.com/science/article/pii/S0958166925000643]
- 8. Next-Generation Sequencing Illumina Workflow–4 Key Steps [https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/next-generation-sequencing/illumina-workflow.html]
- 9. How NGS Automation Improves Library Preparation ... [https://www.euformatics.com/blog-post/how-ngs-automation-improves-library-preparation-efficiency-and-accuracy]
- 10. Challenges in Overcoming with NGS Sample... Workflows Automated [https://dispendix.com/blog/overcoming-challenges-in-ngs-workflows-with-automated-sample-prep]
- 11. High-throughput screening: accelerating drug discovery [https://www.scdiscoveries.com/blog/high-throughput-screening-accelerate-drug-discovery/]
- 12. A Robotic Platform for Quantitative High-Throughput Screening [https://pmc.ncbi.nlm.nih.gov/articles/PMC2651822/]
- 13. High Throughput Screening (HTS) Services [https://www.evotec.com/solutions/drug-discovery-preclinical-development/discovery/hit-identification-services/high-throughput-screening-hts-services]
- 14. High-throughput screening data generation, scoring and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12020062/]
- 15. Color Balance in Illumina Sequencing: Why It Still Matters [https://www.revvity.com/blog/color-balance-illumina-sequencing-why-it-still-matters]
- 16. High-Throughput Variant Detection Using a Color-Mixing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9379672/]
- 17. Automated Methods for NGS Sample Prep [https://sequencing.roche.com/us/en/content/support-for-non-roche-liquid-handlers.html]
- 18. Library prep automation | Automated NGS protocols [https://www.illumina.com/techniques/sequencing/ngs-library-prep/automation.html]
- 19. Next-Generation Sequencing (NGS) [https://dispendix.com/ngs-library-preparation]
- 20. NGS Enrichment Robot; Automated Library Prep Liquid Handling | Agilent [https://www.agilent.com/en/product/automated-liquid-handling/automated-liquid-handling-applications/bravo-ngs]
- 21. Next Generation Sequencing Library Preparation Workstation [https://www.aurorabiomed.com/next-generation-library-sequencing-using-versa-nglp-workstation/?srsltid=AfmBOopNUfClmjfrSJlQYCZcFmZtqyd0J8Lnb941fB-75OxhrzrkpBSd]
- 22. Integrated Lab Automation [https://www.revvity.com/category/integrated-lab-automation]
- 23. Choosing the Right Liquid Handling System for NGS ... [https://dispendix.com/blog/choosing-the-right-liquid-handling-system-for-ngs-applications]
- 24. Integrated DNA Technologies and Beckman Coulter Life Sciences... [https://www.bio-itworld.com/pressreleases/2025/11/13/integrated-dna-technologies-and-beckman-coulter-life-sciences-partner-to-advance-cancer-research-through-automated-ngs-workflows]
- 25. Integrated DNA Technologies and Hamilton Partner to Automate ... [https://www.businesswire.com/news/home/20251001660582/en/Integrated-DNA-Technologies-and-Hamilton-Partner-to-Automate-Customizable-NGS-Workflows-and-Accelerate-Biomarker-Discovery]
- 26. Volta Labs Expands Callisto Automation Through Strategic ... [https://biopharmaapac.com/news/44/7183/volta-labs-expands-callisto-automation-through-strategic-partnership-with-new-england-biolabs.html]
- 27. HP and Tecan Expand Strategic Collaboration to Unveil ... [https://www.hp.com/us-en/newsroom/blogs/2025/HP-Tecan-Expand-Collaboration.html]
- 28. Automated Liquid Handling Makes Next-Generation ... [https://www.bostonind.com/blog/automated-liquid-handling-makes-next-generation-sequencing-more-successful?srsltid=AfmBOordoDL7AiiAtoCC65B9YN3dfO0XTW6s-7AnJKFMFkXVu0XJqD3A]
- 29. Next-generation Sequencing Library Preparation Market ... [https://www.precedenceresearch.com/next-generation-sequencing-library-preparation-market]
- 30. Integration of Acoustic Liquid Handling into Quantitative ... [https://www.sciencedirect.com/science/article/pii/S2472630322010640]
- 31. Accurate Library Preparation in NGS Workflows - Uncoded [https://uncoded.in/the-importance-of-accurate-library-preparation-in-ngs-workflows/]
- 32. 3 Innovations in Next-Generation Sequencing Sample Prep [https://dispendix.com/blog/3-innovations-in-next-generation-sequencing-sample-prep]
- 33. Implementing laboratory automation for next-generation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10373871/]
- 34. 5 Reasons To Automate Your NGS Workflow [https://opentrons.com/archives/resource/5-reasons-to-automate-your-ngs-workflow?srsltid=AfmBOooAIgO97wkJ54T2Voe2cBvBC7Bpv9iONICw-sbqo19-yTdRCDQa]
- 35. NGS is evolving: collaboration and tech lead the way [https://www.drugtargetreview.com/article/168383/ngs-is-evolving-collaboration-and-tech-lead-the-way/]
- 36. The Ultimate Guide to Assay Miniaturization [https://dispendix.com/blog/the-ultimate-guide-to-assay-miniaturization]
- 37. Elevate Your Liquid Handling with Low Volume Non- ... [https://dispendix.com/blog/elevate-your-liquid-handling-with-low-volume-non-contact-dispensers]
- 38. The benefits of non-contact dispensing for fluid control [https://www.news-medical.net/whitepaper/20250417/The-Benefits-of-Non-Contact-Dispensing-for-Fluid-Control.aspx]
- 39. Miniaturization in Genomics, NGS and PCR [In-Depth Guide] [https://www.sptlabtech.com/miniaturization-genomics-ngs-pcr-complete-guide]
- 40. Exploring Miniaturized Reactions Benefits for Research ... [https://dispendix.com/blog/exploring-miniaturized-reactions-benefits-for-research-and-development]
- 41. I.DOT Non Contact Dispenser [https://dispendix.com/idot-non-contact-liquid-handler]
- 42. A Calibration-Free, Noncontact, Disposable Liquid ... [https://www.sciencedirect.com/science/article/pii/S2472630322015540]
- 43. Next-Generation Sequencing ( NGS ) Services in the Real World... [https://www.linkedin.com/pulse/next-generation-sequencing-ngs-services-real-oa6bf]
- 44. The Need for NGS automation for single-cell and spatial ... [https://www.revvity.com/blog/need-ngs-automation]
- 45. Homepage - Clear Labs [https://www.clearlabs.com/]
- 46. Clear Labs Showcases Oncology Initiatives and... NGS Automation [https://www.prweb.com/releases/clear-labs-showcases-oncology-ngs-automation-initiatives-and-announces-collaboration-with-labcorp-302615804.html]
- 47. NGS Quantification, Normalization & Pooling | Myra ... [https://biomolecularsystems.com/applications/ngs-quantification-normalisation-and-pooling/]
- 48. NGS Data Analysis for Illumina Platform—Overview and ... [https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/next-generation-sequencing/ngs-data-analysis-illumina.html]
- 49. CSI NGS Portal: An Online Platform for Automated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7312552/]
- 50. How-to: NGS Quality Control [https://frontlinegenomics.com/how-to-ngs-quality-control/]
- 51. Statistical guidelines for quality control of next-generation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8408346/]
- 52. Integrating Artificial Intelligence in Next-Generation Sequencing [https://pmc.ncbi.nlm.nih.gov/articles/PMC12191491/]
- 53. The Power of NGS Library Prep Automation: Enhancing ... [https://dispendix.com/blog/power-of-ngs-library-prep-automation-enhancing-genomics-with-automated-liquid-handling-and-dna-clean-up]
- 54. 2025 and Beyond: The Future of Genomic Data Analysis ... [https://blog.crownbio.com/2025-and-beyond-the-future-of-genomic-data-analysis-and-innovations-in-genomics-services]
- 55. Rapid NGS Analysis on Google Cloud Platform [https://pmc.ncbi.nlm.nih.gov/articles/PMC12627752/]
- 56. Automated NGS Workflows for Biomarker Discovery | IDT [https://sg.idtdna.com/pages/about/news/2025/10/01/integrated-dna-technologies-and-hamilton-company-partner-to-automate-customizable-ngs-workflows-and-accelerate-biomarker-discovery]
- 57. IDT & Beckman Coulter Partner to Automate NGS Workflows [https://lifesciences.danaher.com/us/en/news/automated-ngs-workflows-cancer-research]
- 58. Genomics Cloud Computing | Technology from Bluebee & ... [https://www.illumina.com/science/technology/development/genomics-cloud-computing.html]
- 59. A deep learning framework for high-throughput mechanism ... [https://www.nature.com/articles/s42256-020-00285-9]
- 60. Basepair: NGS Data Analysis Platform [https://www.basepairtech.com/]
- 61. The Next-Generation Sequencing Quality Initiative and ... [https://wwwnc.cdc.gov/eid/article/31/13/24-1175_article]
- 62. Standardization and quality management in next- ... [https://www.sciencedirect.com/science/article/pii/S2212066116300230]
- 63. Standardizing Clinical NGS Applications—Based on ... [https://www.cambridgepublish.com/csa/article/view/155]
- 64. An NGS Workflow Blueprint for DNA Sequencing Data and Its ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4827795/]
- 65. Automated NGS Library Preparation System Market 2025- ... [https://www.360iresearch.com/library/intelligence/automated-ngs-library-preparation-system]
- 66. NGS Data Analysis: Best Practices and Common Pitfalls [https://dromicsedu.com/blog-details/ngs-data-analysis-best-practices-and-common-pitfalls]
- 67. Adopting NGS Technology for Routine Genetic... | Today's Clinical Lab [https://www.clinicallab.com/adopting-ngs-technology-for-routine-genetic-profiling-26230]
- 68. A rapid turnaround for a cytological liquid biopsy assay... time workflow [https://pubmed.ncbi.nlm.nih.gov/39704279/]
- 69. How to automate NGS library preparation [https://frontlinegenomics.com/how-to-automate-ngs-library-preparation/]
- 70. The Complete Guide to Automated Sample Preparation for ... [https://dispendix.com/blog/the-complete-guide-to-automated-sample-preparation-for-ngs]
- 71. Validation between manual and automated NGS library prep [https://www.aurorabiomed.com/validation-of-verogens-ngs-forenseq-workflow-kintelligence-kit-mtdna-whole-genome-kit-signature-prep-kit/?srsltid=AfmBOoqKIXqpzTjeTuJ7-a4Z7S0X6Lf1uBHoIdsg_YxXTlG0zp7wVZSM]
- 72. How to increase efficiency in Routine Oncology NGS Workflows [https://www.thermofisher.com/ru/ru/home/events/increase-efficiency-routine-oncology-ngs-workflows-webinar.html]
- 73. Common NGS Pitfalls of CLIA Certification and ISO ... [https://a2la.org/common-ngs-pitfalls-of-clia-certification-and-iso-accreditation/]
- 74. The New EU Regulation on In Vitro Diagnostic Medical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8061679/]
- 75. In Vitro Diagnostic Regulation (IVDR)—Frequently Asked ... [https://www.thermofisher.com/us/en/home/clinical/ivdr/faqs.html]
- 76. Automated capture-based NGS workflow: one thousand ... [https://pubmed.ncbi.nlm.nih.gov/34142509/]
- 77. Automating NGS [https://insights.opentrons.com/ngs-automation]
- 78. Go ahead and peek! Technical documentation under the IVDR & MDR [https://www.seleon.com/en/regulatory-affairs/go-ahead-and-peek-technical-documentation-under-the-ivdr-mdr/]
- 79. DNA Sequencing Technologies Compared: Illumina ... [https://synapse.patsnap.com/article/dna-sequencing-technologies-compared-illumina-nanopore-and-pacbio]
- 80. A Primer on NGS Technologies and Their Specs (2025) [https://blog.latch.bio/p/a-primer-on-ngs-technologies-and]
- 81. Comparing long-read sequencing technologies [https://www.pacb.com/blog/sequencing-101-comparing-long-read-sequencing-technologies/]
- 82. Automated NGS Workflows for Cancer Research | IDT [https://eu.idtdna.com/pages/about/news/2025/11/12/integrated-dna-technologies-and-beckman-coulter-life-sciences-partner-to-advance-cancer-research-through-automated-ngs-workflows]
- 83. Automation of customizable library preparation for next- ... [https://www.nature.com/articles/s41598-024-67950-6]
- 84. Comparative analysis of Illumina, PacBio, and nanopore ... [https://www.frontiersin.org/journals/microbiomes/articles/10.3389/frmbi.2025.1587712/full]
- 85. NGS Library Preparation Automation Market [https://www.researchandmarkets.com/reports/5925147/ngs-library-preparation-automation-market?srsltid=AfmBOorlI6gnYVvgxO1Y_okRIB8BrtwXRk5lHHIeR2ZsAiXC-k0O08GK]
- 86. Lab Automation Market Growth, Drivers, and Opportunities [https://www.marketsandmarkets.com/Market-Reports/lab-automation-market-1158.html]
- 87. Next Generation Sequencing (NGS) Market Poised to Hit ... [https://www.prnewswire.com/news-releases/next-generation-sequencing-ngs-market-poised-to-hit-42-25-billion-by-2033-key-drivers-behind-the-surge--latest-study-by-grand-view-research-inc-302614298.html]
- 88. Considerations for the Automation of Sequencing Workflows [https://formulatrix.com/life-science-automation-blog/considerations-for-the-automation-of-sequencing-workflows/]
- 89. Global High-Throughput Sequencing NGS Market 2025-2033 [https://www.linkedin.com/pulse/global-high-throughput-sequencing-ngs-market-9owae/]
- 90. Next Generation Sequencers Market Forecast Report 2025 ... [https://finance.yahoo.com/news/next-generation-sequencers-market-forecast-080200693.html]
- 91. maximizing return on investment through effective life-cycle ... [https://pubmed.ncbi.nlm.nih.gov/21906566/]