Strategies for Reducing Host DNA Background in Metagenomic NGS: A Comprehensive Guide for Researchers
High host DNA background remains a significant challenge in metagenomic next-generation sequencing (mNGS), particularly in clinical samples like blood, respiratory secretions, and tissues where host content can exceed 99%.
Strategies for Reducing Host DNA Background in Metagenomic NGS: A Comprehensive Guide for Researchers
Abstract
High host DNA background remains a significant challenge in metagenomic next-generation sequencing (mNGS), particularly in clinical samples like blood, respiratory secretions, and tissues where host content can exceed 99%. This comprehensive review explores current methodologies for host DNA depletion, comparing physical, enzymatic, and bioinformatic approaches. We examine the impact of host depletion on diagnostic sensitivity, microbial read enrichment, and community representation across diverse sample types. Recent advancements including novel filtration technologies and optimized DNA extraction methods are evaluated for their efficacy in improving pathogen detection while preserving microbial integrity. This article provides researchers and clinicians with evidence-based guidance for selecting appropriate host depletion strategies to enhance mNGS performance in infectious disease diagnostics and microbiome studies.
The Host DNA Challenge: Understanding the Fundamental Barrier in Metagenomic NGS
The Critical Impact of Host DNA on Sequencing Efficiency and Sensitivity
FAQs: Understanding Host DNA in NGS
Q1: Why is host DNA a significant problem in metagenomic next-generation sequencing (mNGS)? Host DNA is a major problem because it consumes the vast majority of sequencing reads, leaving limited capacity for detecting microbial pathogens. In samples like blood and respiratory secretions, host DNA can constitute over 99% of the total sequenced DNA [1] [2]. This overwhelming background leads to reduced sensitivity for identifying low-abundance microbes and significantly increases the cost and depth of sequencing required to obtain meaningful microbial data [3] [4].
Q2: Which sample types are most affected by high host DNA content? The proportion of host DNA varies significantly by sample type [3]:
- High host DNA (>90%): Blood, bronchoalveolar lavage (BAL), sputum, saliva, and nasopharyngeal swabs [1] [2] [4].
- Low host DNA (<10%): Stool samples [3].
Q3: What is the relationship between host DNA content and sequencing depth? As host DNA content increases, the required sequencing depth to achieve sufficient microbial genome coverage increases exponentially. Studies have shown that in samples with 90% host DNA, a reduction in sequencing depth majorly impacts sensitivity, increasing the number of undetected microbial species. Even with a fixed depth of 10 million reads, microbiome profiling becomes increasingly inaccurate as host DNA levels rise [3] [4].
Q4: Can I use bioinformatics to remove host DNA sequences? Yes, bioinformatic tools like KneadData (which uses Bowtie2) can map and remove reads that align to the host genome after sequencing [3] [4]. However, this is a post-sequencing corrective measure. It does not solve the fundamental problem of wasted sequencing resources on non-informative host reads, making pre-sequencing host depletion a more efficient strategy for enriching microbial signals [1].
Troubleshooting Guides
Problem: Low Microbial Read Counts in mNGS
Possible Causes & Solutions:
| Problem Area | Specific Issue | Recommended Solution |
|---|---|---|
| Sample Type | Using high-host content samples (e.g., blood, BAL) without depletion. | Implement a pre-sequencing host depletion method tailored to your sample type [1] [2]. |
| Host Depletion Method | Method is inefficient, labor-intensive, or alters microbial composition. | Evaluate advanced methods like the ZISC-based filtration, which showed >99% WBC removal without affecting microbial integrity [1] [5]. |
| DNA Input | Using cell-free DNA (cfDNA) from plasma for septic samples. | For sepsis, use genomic DNA (gDNA) from cell pellets combined with host cell depletion. One study showed gDNA-based mNGS detected pathogens in 100% of samples, outperforming cfDNA-based methods [1]. |
| Sequencing Depth | Inadequate sequencing depth for the level of host DNA contamination. | Increase sequencing depth significantly for samples with >90% host DNA. For context, one clinical study sequenced at least 10 million reads per sample on a NovaSeq 6000 [1] [3]. |
Problem: Contamination in mNGS Workflow
Possible Causes & Solutions:
| Problem Area | Specific Issue | Recommended Solution |
|---|---|---|
| Lab Layout | Pre- and post-PCR areas are not physically separated. | Designate and use distinct areas for sample preparation, PCR setup, and post-PCR analysis. Restrict equipment (pipettes, lab coats) to these dedicated areas [6]. |
| Reagents | Reagents are contaminated or cross-used. | Prepare and store reagents separately. Aliquot reagents in small portions designated for pre- or post-PCR use only [6]. |
| Practice | Amplicons from previous runs contaminate new reactions. | Always use pipette tips with aerosol filters. Never bring reagents or equipment from a post-PCR area back to a pre-PCR area [6]. |
| Controls | Contamination is not detected early. | ALWAYS include a negative control reaction (using ultrapure water instead of template DNA) in every run to check for contamination [6]. |
Experimental Protocols & Data
Detailed Methodology: ZISC-Based Filtration for Host Depletion in Blood
This protocol is adapted from a 2025 study that optimized mNGS for sepsis diagnosis [1].
1. Sample Preparation:
- Collect whole blood sample (e.g., 4 mL) in an appropriate anticoagulant tube.
- Optionally, spike with an internal reference control (e.g., ZymoBIOMICS Spike-in Control) to monitor microbial recovery.
2. Host Cell Depletion Filtration:
- Secure the novel ZISC-based fractionation filter (e.g., Devin filter from Micronbrane) onto a syringe.
- Transfer the blood sample into the syringe.
- Gently depress the plunger to push the blood through the filter into a clean collection tube (e.g., 15 mL Falcon tube).
- The filter achieves >99% white blood cell (WBC) removal while allowing bacteria and viruses to pass through unimpeded.
3. Plasma and Cell Pellet Separation:
- Centrifuge the filtered blood at low speed (e.g., 400g for 15 minutes) to isolate the plasma.
- Transfer the plasma to a new tube and perform high-speed centrifugation (e.g., 16,000g) to obtain a microbial cell pellet.
4. DNA Extraction:
- Extract genomic DNA (gDNA) from the microbial cell pellet using a dedicated microbial DNA enrichment kit.
- For comparison, cell-free DNA (cfDNA) can be extracted from the plasma supernatant.
5. Library Preparation and Sequencing:
- Prepare mNGS libraries using an Ultra-Low Library Prep Kit.
- Sequence on a platform such as Illumina NovaSeq 6000, aiming for a minimum of 10 million reads per sample.
Quantitative Data on Host Depletion Efficacy
Table 1: Comparison of Host Depletion Methods on Clinical Samples [1]
| Method | Principle | Host Depletion Efficiency | Key Findings in Clinical Sepsis Samples |
|---|---|---|---|
| Novel ZISC Filtration | Physical retention of host WBCs via a zwitterionic coating. | >99% WBC removal [1]. | mNGS with filtered gDNA detected all expected pathogens in 100% (8/8) of samples, with an average of 9351 microbial RPM (reads per million)—a tenfold increase over unfiltered samples (925 RPM) [1]. |
| Differential Lysis (QIAamp DNA Microbiome Kit) | Selective lysis of human cells. | Varies by sample type [2]. | More labor-intensive; efficiency lower than novel filtration in side-by-side comparison [1]. |
| Methylated DNA Removal (NEBNext Microbiome DNA Enrichment Kit) | Removal of CpG-methylated host DNA. | Varies by sample type [2]. | Preserved microbial reads but was less efficient than novel filtration [1]. |
Table 2: Impact of Host DNA Percentage and Sequencing Depth on Microbial Detection [3] [4]
| Host DNA in Sample | Sequencing Depth | Impact on Microbial Detection Sensitivity |
|---|---|---|
| 10% | Standard Depth (e.g., 5-10 M reads) | Good sensitivity for most species. |
| 90% | Standard Depth | Decreased sensitivity; increased number of undetected species, particularly low-abundance ones. |
| 90% | Reduced Depth | Major impact on sensitivity; significant loss of microbial species information. |
| 99% | Fixed Depth of 10 M reads | Profiling becomes highly inaccurate due to extremely low effective microbial depth. |
Workflow Diagrams
Host Depletion mNGS Workflow
Host Depletion Decision Pathway
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Key Reagents and Kits for Host DNA Depletion
| Product Name | Manufacturer | Principle / Function | Key Application Note |
|---|---|---|---|
| Devin Filter (ZISC-based) | Micronbrane | Zwitterionic coating binds and retains host leukocytes physically. | Achieved >99% WBC removal from blood; enabled 10x enrichment of microbial reads in mNGS [1]. |
| QIAamp DNA Microbiome Kit | Qiagen | Differential lysis of human cells to enrich for microbial DNA. | One of several methods evaluated for respiratory samples; performance varies by sample matrix [1] [2]. |
| NEBNext Microbiome DNA Enrichment Kit | New England Biolabs | Binds and removes CpG-methylated host DNA, enriching non-methylated microbial DNA. | A post-extraction method compared favorably but was less efficient than novel filtration in one study [1]. |
| HostZERO Microbial DNA Kit | Zymo Research | Chemical and enzymatic degradation of host DNA. | Effectively decreased host DNA in frozen nasal (73.6% decrease) and sputum samples [2]. |
| MolYsis Kit | Molzym | Selective lysis of human cells and degradation of released DNA. | Effective for sputum, decreasing host DNA by 69.6% [2]. |
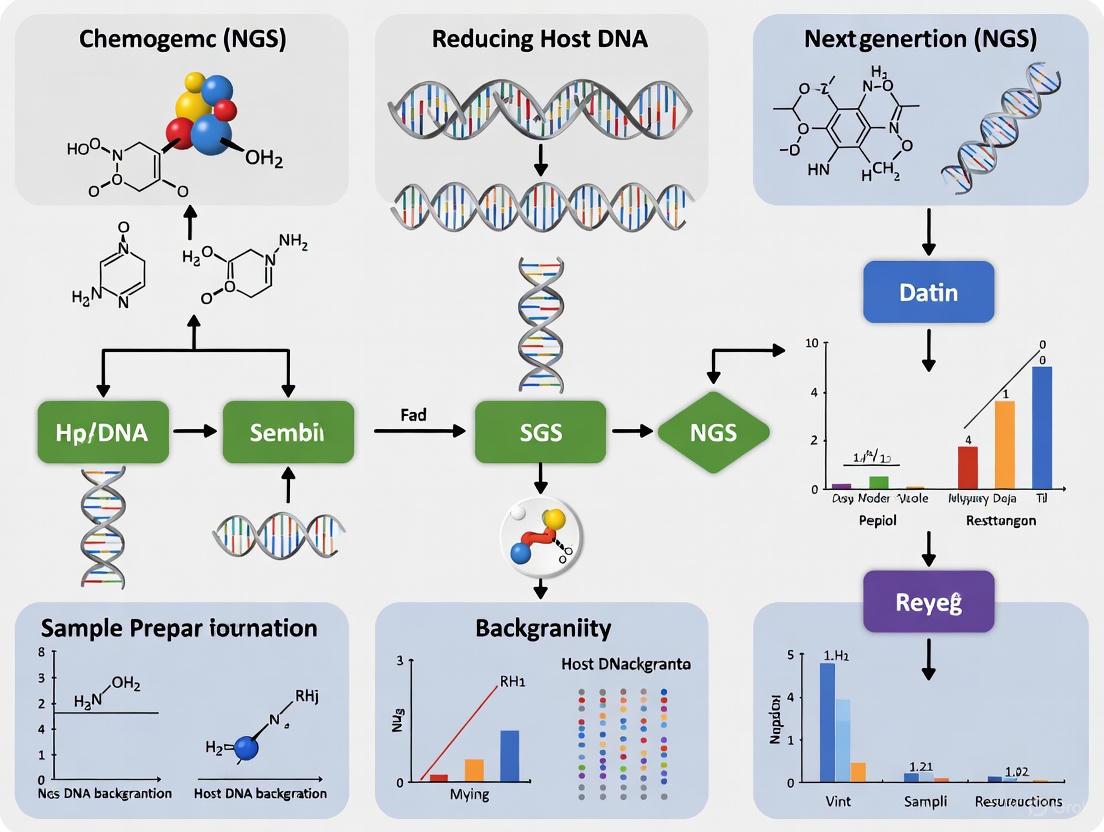
In chemogenomic Next-Generation Sequencing (NGS) research, host DNA contamination represents a significant bottleneck that can compromise data quality and experimental outcomes. Excessive host DNA in samples reduces microbial or target pathogen sequencing depth, increases sequencing costs, and can obscure genuine biological signals. This technical guide addresses the critical need for accurate quantification of host DNA proportions and provides evidence-based strategies for effective host depletion, enabling more sensitive and accurate NGS results in drug discovery and development workflows.
Quantitative Analysis of Host DNA Background
The proportion of host DNA varies significantly across different sample types, directly impacting the effectiveness of downstream NGS applications. The following table summarizes documented host DNA levels across various clinical and experimental samples:
Table 1: Host DNA Proportions Across Sample Types
| Sample Type | Host DNA Proportion | Post-Treatment Host DNA | Enhancement Method | Key Findings |
|---|---|---|---|---|
| Blood (sepsis patients, gDNA-based mNGS) | High background (average 925 RPM microbial reads) | >10x increase in microbial reads (9351 RPM) | Novel ZISC-based filtration [5] | >99% white blood cell removal; significantly improved pathogen detection [5] |
| Swab specimens (COVID-19 patients) | Variable (impacted SARS-CoV-2 detection sensitivity) | Improved detection rate (92.9% for Ct ≤35) | Host DNA removal via DNA enzyme digestion [7] | Host removal enhanced sensitivity without affecting microbial RNA abundance [7] |
| Bacterial WGS from pure cultures | Contamination present in multiple studies | Taxonomic filtering enabled accurate variant calling | Kraken-based taxonomic classification [8] | 45% of samples in some studies had <90% reads from target organism [8] |
| Therapeutic proteins (CHO host cells) | Residual host cell DNA impurity | Detection limit of 0.1-0.8 ppb | Direct qPCR without DNA extraction [9] | Proteinase K/SDS digestion with Tween 20 to prevent inhibition [9] |
Host DNA Quantification Methods
Accurate quantification of host DNA is essential for assessing sample quality and determining the need for host depletion procedures. The following methods are commonly employed:
Table 2: Host DNA Quantification Methods
| Method | Principle | Sensitivity | Advantages | Limitations |
|---|---|---|---|---|
| UV Absorbance [10] [11] | Measures absorbance at 260nm | Limited sensitivity at low concentrations | Quick, simple, no special reagents | Cannot distinguish between DNA and RNA [10] |
| Fluorescence Dyes (PicoGreen, SYBR Green) [10] [11] | Fluorescent dyes bind dsDNA | High sensitivity for low concentrations | Specific for dsDNA, more sensitive than UV | Requires standard curve, dye-specific [10] |
| qPCR/dPCR [9] [12] | Target-specific amplification | Very high (detection to 0.1 ppb) | Host-specific, extremely sensitive | Requires host-specific primers/probes [9] |
| Capillary Electrophoresis [10] [11] | Size separation with fluorescence | Moderate | Fragment size distribution, automated | Equipment intensive, lower throughput [10] |
Experimental Protocols for Host DNA Assessment and Depletion
Protocol 1: Direct qPCR for Residual Host Cell DNA Quantification
This protocol enables precise quantification of host DNA without extraction steps, adapted from Peper et al. [9]:
- Protein Digestion: Mix sample with proteinase K and SDS to digest therapeutic proteins and release DNA.
- Add Stabilizers: Include Tween 20 and NaCl to minimize precipitation of therapeutic proteins during digestion.
- qPCR Setup: Prepare qPCR mix with 2% (v/v) Tween 20 to counteract SDS inhibition.
- Amplification: Use host-specific primers and probes (e.g., CHO-specific for mammalian cell lines).
- Quantification: Calculate residual DNA against standard curve, with LOD of 0.1-0.8 ppb for most proteins.
This method has been validated according to ICH guidelines and applied to 25 different therapeutic proteins [9].
Protocol 2: Taxonomic Filtering for Bacterial WGS Contamination Removal
For comprehensive removal of contaminant reads in bacterial whole-genome sequencing:
- Taxonomic Classification: Process sequencing reads with Kraken classifier against curated database.
- Filter Implementation: Remove all reads not classified to target genus (e.g., non-Acinetobacter reads for A. baumannii studies).
- Variant Calling: Perform standard mapping and SNP calling on filtered read set.
- Validation: Compare variant profiles before and after filtering to assess contamination impact.
This approach has been shown to eliminate hundreds of false positive and negative SNPs even in slightly contaminated samples [8].
Protocol 3: Host DNA-Removed Metagenomic Sequencing
For swab and clinical specimens requiring pathogen detection:
- Nucleic Acid Extraction: Extract total nucleic acid using automated system (e.g., 200μl sample input).
- Host DNA Depletion: Treat with DNA enzyme and buffer to digest DNA and enrich for RNA pathogens.
- Reverse Transcription: Synthesize cDNA from remaining RNA.
- Library Preparation: Use probe-anchored polymer sequencing method for library construction.
- Sequencing and Analysis: Sequence on appropriate platform (e.g., MGSEQ-2000) and remove any residual human reads bioinformatically.
This workflow achieved 92.9% detection rate for SARS-CoV-2 in samples with Ct values ≤35 [7].
Workflow Visualization: Host DNA Management in NGS
Research Reagent Solutions
Table 3: Essential Reagents for Host DNA Management
| Reagent/Kit | Function | Application Note |
|---|---|---|
| Ribo-Zero [13] | rRNA depletion | Reduces rRNA to <1%, maintains transcript representation |
| Proteinase K with SDS [9] | Protein digestion for direct qPCR | Enables residual DNA detection without extraction |
| Kraken Classifier [8] | Taxonomic read classification | Filters contaminant reads at genus/species level |
| ZISC-based Filtration [5] | Physical host cell depletion | >99% WBC removal while preserving microbes |
| PicoGreen/SYBR Green [10] [11] | dsDNA quantification | Fluorometric detection specific to dsDNA |
| DNA Enzyme Treatment [7] | Selective host DNA removal | Digests DNA while preserving RNA pathogens |
| qPCR Host-Specific Primers [9] [12] | Targeted host DNA detection | Enables sensitive residual DNA quantification |
Frequently Asked Questions
Q1: What is the acceptable threshold for host DNA proportion in NGS samples? The acceptable threshold varies by application. For metagenomic sequencing aiming to detect low-abundance pathogens, host DNA should ideally be reduced to <80% of total reads. Studies show that novel filtration methods can achieve >99% host cell removal, resulting in over tenfold increase in microbial reads [5].
Q2: Can I use UV spectrophotometry (A260/A280) alone to assess host DNA contamination? While UV spectrophotometry provides rapid assessment of nucleic acid purity (ideal A260/A280 ratio of 1.8-2.0 for DNA), it cannot distinguish between host and target DNA, has limited sensitivity at low concentrations, and may miss contamination that doesn't affect the absorbance ratio [10]. For host-specific quantification, qPCR with host-specific primers is recommended [9] [12].
Q3: What is the most effective host depletion method for blood samples? For blood samples, the novel ZISC-based filtration device has demonstrated excellent performance with >99% white blood cell removal across various blood volumes while allowing unimpeded passage of bacteria and viruses. This method achieved an average of 9351 microbial RPM compared to 925 RPM in unfiltered samples in sepsis patient testing [5].
Q4: How does host DNA removal affect the representation of the microbial community? Properly implemented host DNA removal methods specifically target host nucleic acids while preserving microbial composition. Studies comparing workflows with and without host removal found that effective host depletion does not alter the microbial composition, making it suitable for accurate pathogen profiling [5] [7].
Q5: What bioinformatic approaches can help address host DNA contamination? Taxonomic classification tools like Kraken can filter contaminant reads bioinformatically. This approach has been shown to remove hundreds of false positive and negative SNPs even in slightly contaminated samples. For comprehensive contamination removal, combine wet-lab depletion with bioinformatic filtering [8].
Frequently Asked Questions (FAQs)
Q1: What is the core problem with host DNA in microbial NGS?
Host DNA acts as a major contaminant in metagenomic next-generation sequencing (mNGS). In samples derived from human hosts (e.g., tissues, blood, saliva), the human genome can constitute over 90% of the total DNA sequenced [14] [15]. This overwhelms the microbial signal, leading to two critical issues:
- Obscured Detection: The vast majority of sequencing reads are consumed by host DNA, drastically reducing the number of reads available to identify pathogens or microbiome members, thereby lowering detection sensitivity [3] [16].
- Wasted Resources: Sequencing depth is wasted on uninformative host reads, increasing costs and computational burden without yielding useful microbial data [17] [15].
Q2: Which sample types are most affected by this issue?
The impact of host DNA varies significantly by sample type, primarily due to differences in the microbial-to-host cell ratio.
- High-Host-DNA Samples ( >90% human reads): Saliva, skin swabs, nasal swabs, vaginal swabs, bronchoalveolar lavage fluid (BALF), and biopsy tissues [14] [15].
- Low-Host-DNA Samples ( <10% human reads): Stool samples [3] [15].
Q3: My 16S rRNA sequencing of colon biopsies shows strange results. Could host DNA be the cause?
Yes. During 16S amplicon sequencing, PCR primers can mis-prime, or mistakenly bind, to similar sequences in the host genome. This generates "host off-target" sequences that are misclassified as bacterial [17]. This is a significant issue with the commonly used V3-V4 primers, where mis-priming to human chromosomes 5, 11, and 17 can lead to false bacterial identifications and obscure true differences in microbiota composition [17].
Q4: What are the main strategies to deplete host DNA?
Strategies can be applied either before DNA extraction ("pre-extraction") or after DNA extraction ("post-extraction").
Pre-extraction Methods: These leverage physical or chemical differences between host and microbial cells.
- Selective Lysis: Using mild detergents (e.g., saponin) or osmotic lysis to break open fragile human cells while leaving robust microbial cells with cell walls intact [14] [15].
- Nuclease Treatment: Adding DNase enzymes to degrade the exposed host DNA after lysis. The intact microbes are then purified for DNA extraction [18] [15].
- Propidium Monoazide (PMA) Treatment: Following selective lysis, PMA dye penetrates damaged host cells, binds to DNA, and upon light exposure, permanently cross-links it, preventing its amplification in downstream steps [14].
Post-extraction Methods: These exploit genomic differences.
- Methylation-Based Capture: Eukaryotic DNA is heavily methylated. Kits using methyl-binding domain (MBD) proteins can capture and remove this methylated host DNA, leaving behind relatively unmethylated microbial DNA [15].
Troubleshooting Guides
Problem: Low Sensitivity for Microbial Pathogens in BALF Samples
Potential Cause: Bronchoalveolar lavage fluid (BALF) samples are typically dominated by host DNA (>95%), which can mask the signal from intracellular pathogens like Mycobacterium tuberculosis [16].
Solutions:
- Implement a host depletion protocol. A saponin-based host DNA depletion assisted (HDA) method has been shown to significantly improve results.
- Follow this optimized HDA-mNGS protocol for BALF [16]:
- Sample Pre-treatment: Add Sputasol to the BALF sample and incubate at room temperature for 2-5 minutes.
- Centrifugation: Pellet the microbial cells.
- Selective Lysis & DNase Treatment: Resuspend the pellet and lyse the cells. Then, treat with a salt-active nuclease (e.g., HL-SAN) which is highly effective at degrading host DNA under high-salt conditions [18].
- DNA Extraction & Sequencing: Proceed with DNA extraction using a kit like the TIANamp Micro DNA Kit, followed by library preparation and sequencing.
- Expected Outcome: This method increased the sensitivity for diagnosing pulmonary tuberculosis from 51.2% (conventional mNGS) to 72.0% and provided up to a 16-fold increase in MTB genome coverage [16].
Problem: High Host DNA Background in Saliva Metagenomics
Potential Cause: Saliva contains large amounts of human epithelial cells and extracellular host DNA, routinely resulting in >90% human sequencing reads [14].
Solutions:
- Apply a simple osmotic lysis and PMA (lyPMA) treatment.
- Detailed lyPMA Protocol [14]:
- Resuspend the saliva pellet in pure water to osmotically lyse mammalian cells.
- Add PMA to a final concentration of 10 µM and incubate in the dark for 5 minutes.
- Place the sample on ice and expose it to a 650-W halogen light source for 2 minutes to photo-activate the PMA.
- Proceed with standard DNA extraction.
- Expected Outcome: This cost-effective method reduced host-derived sequencing reads from 89.29% in untreated samples to 8.53%, with minimal taxonomic bias [14].
The following table quantifies how increasing levels of host DNA reduce the sensitivity of Whole Metagenome Sequencing (WMS) for detecting microbial species.
Table 1: Impact of Host DNA Proportion and Sequencing Depth on Microbial Detection Sensitivity in WMS [3]
| Proportion of Host DNA | Sequencing Depth | Key Impact on Microbial Profiling |
|---|---|---|
| 10% | Variable | Minimal impact; high sensitivity for most species. |
| 90% | Standard Depth (~5-10M reads) | Decreased sensitivity; failure to detect very low and low-abundance species. |
| 90% | Reduced Depth | Major impact; significant increase in the number of undetected species. |
| 99% | Fixed Depth (10M reads) | Highly inaccurate and incomplete profiling due to insufficient microbial reads. |
Table 2: Comparison of Host DNA Depletion Methods
| Method | Principle | Best For | Advantages | Limitations |
|---|---|---|---|---|
| Osmotic Lysis + PMA (lyPMA) [14] | Selective lysis of host cells followed by photo-induced cross-linking of free DNA. | Fresh or frozen saliva, other host-derived samples. | Cost-effective, rapid (<5 min hands-on), low taxonomic bias. | Optimized for specific sample types. |
| Selective Lysis + Salt-Active Nuclease (e.g., HL-SAN) [16] [18] | Selective lysis followed by enzymatic degradation of host DNA in high-salt buffers. | BALF, sputum, wound swabs (targeting robust pathogens). | Highly efficient (1000-fold reduction in host DNA), robust, proven in clinical workflows. | High salt conditions may not be suitable for fragile enveloped viruses. |
| Methylation-Based Depletion [15] | Binding and removal of methylated eukaryotic DNA with MBD-bound beads. | Various samples where microbial DNA is largely unmethylated. | Post-extraction method; does not require intact cells. | Bias against microbes with methylated genomes or AT-rich genomes [14]. |
Workflow Diagrams
The following diagram illustrates the logical decision process for selecting a host DNA depletion strategy.
Diagram 1: Decision Workflow for Host DNA Depletion Strategy Selection
This diagram outlines the general workflow for the pre-extraction host DNA depletion method using selective lysis and nuclease treatment.
Diagram 2: Pre-extraction Host DNA Depletion Workflow
The Scientist's Toolkit: Key Reagents for Host DNA Depletion
Table 3: Essential Reagents for Host DNA Depletion Protocols
| Reagent / Kit | Function / Principle | Specific Example(s) |
|---|---|---|
| Saponin | A non-ionic detergent for selective lysis of mammalian cell membranes without disrupting microbial cell walls [15]. | Used in HDA-mNGS protocol for BALF samples [16]. |
| Salt-Active Nuclease (HL-SAN) | A nuclease that achieves optimal activity under high-salt conditions, effectively degrading host DNA after lysis. | ArcticZymes HL-SAN; used in multiple clinical metagenomic studies [16] [18]. |
| Propidium Monoazide (PMA) | A DNA intercalating dye that penetrates only membrane-compromised cells. Upon light exposure, it covalently cross-links DNA, blocking PCR amplification [14]. | Used in the lyPMA protocol for saliva samples [14]. |
| Methyl-Binding Domain (MBD) Kits | Post-extraction method that uses MBD proteins bound to magnetic beads to capture and remove methylated host DNA. | NEBNext Microbiome DNA Enrichment Kit [14] [15]. |
Frequently Asked Questions (FAQs)
Q1: Why is host DNA background a major problem in chemogenomic NGS studies of pathogens? The overwhelming abundance of host DNA in samples consumes the majority of sequencing capacity, leaving few reads for detecting pathogenic organisms. In blood samples, the high concentration of human DNA can severely limit the sensitivity of metagenomic Next-Generation Sequencing (mNGS) for pathogen detection [5] [19].
Q2: What are the main methods to reduce host DNA background? There are two primary approaches: (1) Pre-extraction methods that physically remove host cells (e.g., white blood cells) before DNA extraction, using techniques like differential lysis or novel filtration devices, and (2) Post-extraction methods that selectively remove or deplete host DNA after extraction, for example, by exploiting differences in DNA methylation patterns [5] [19].
Q3: How does whole-cell DNA (wcDNA) mNGS compare to cell-free DNA (cfDNA) mNGS for pathogen detection? wcDNA mNGS demonstrates significantly higher sensitivity for pathogen detection in clinical body fluid samples. One study reported a concordance rate with culture results of 63.33% for wcDNA mNGS versus 46.67% for cfDNA mNGS [20]. Furthermore, the mean proportion of host DNA in wcDNA mNGS (84%) was significantly lower than in cfDNA mNGS (95%) [20].
Q4: What are common sequencing preparation failures and their causes? Common issues include low library yield, adapter contamination, and over-amplification artifacts. Root causes often involve poor input DNA/RNA quality, contaminants inhibiting enzymes, inaccurate quantification, inefficient adapter ligation, or overly aggressive purification leading to sample loss [21].
Troubleshooting Guides
Problem: Low Microbial Read Counts Due to High Host DNA Background
| Observed Symptom | Potential Cause | Diagnostic Check | Corrective Action |
|---|---|---|---|
| Low percentage of microbial reads despite high total sequencing reads. | Inefficient host cell depletion. | Check pre-filtration and post-filtration cell counts; assess host DNA percentage in sequenced data. | Implement a robust host depletion method, such as the ZISC-based filtration, which can achieve >99% white blood cell removal [5] [19]. |
| Inconsistent pathogen detection sensitivity. | Reliance on cell-free DNA (cfDNA). | Compare microbial read counts from cfDNA vs. whole-cell DNA (wcDNA) from cell pellets. | Switch to a gDNA-based mNGS workflow from cell pellets, which is more effectively enhanced by host depletion methods [19]. |
| High host DNA percentage in wcDNA mNGS. | Suboptimal sample processing. | Review centrifugation protocols for cell pellet preparation. | Optimize the centrifugation steps to ensure effective separation of microbial cells from host components in the sample [20]. |
Problem: General NGS Library Preparation Failures
| Observed Symptom | Potential Cause | Diagnostic Check | Corrective Action |
|---|---|---|---|
| Low library yield. | Poor input quality or contaminants (e.g., phenol, salts). | Check nucleic acid purity via spectrophotometry (A260/A280 and A260/230 ratios). A ratio of ~1.8 is desirable for DNA [22]. Re-purify input sample; use fluorometric quantification (e.g., Qubit) instead of UV absorbance alone [21]. | |
| Adapter-dimer contamination (sharp peak ~70-90 bp). | Suboptimal adapter ligation conditions; inefficient purification. | Analyze library profile using an instrument like BioAnalyzer or TapeStation [21]. | Titrate adapter-to-insert molar ratio; optimize bead-based cleanup parameters to remove short fragments [21]. |
| Over-amplification artifacts; high duplication rate. | Too many PCR cycles during library amplification. | Review library amplification protocol and cycle number. | Reduce the number of PCR cycles; amplify from leftover ligation product rather than over-cycling a weak product [21]. |
Experimental Protocols & Data
This protocol details a novel pre-extraction method to deplete host white blood cells.
- Sample Preparation: Collect whole blood using standard phlebotomy techniques. Blood samples should be processed fresh for optimal results.
- Filtration Setup: Securely connect the novel ZISC-based fractionation filter (e.g., Devin filter) to a syringe.
- Host Cell Depletion: Transfer approximately 4 mL of whole blood into the syringe. Gently depress the plunger to push the blood sample through the filter into a clean collection tube. This step removes >99% of white blood cells while allowing bacteria and viruses to pass through unimpeded.
- Centrifugation: Centrifuge the filtered blood at low speed (e.g., 400g for 15 min) to isolate plasma. Subject the plasma to high-speed centrifugation (e.g., 16,000g) to obtain a microbial cell pellet.
- DNA Extraction: Proceed with microbial DNA extraction from the pellet using a commercial kit.
This protocol allows researchers to compare the performance of two primary mNGS approaches.
- Sample Processing: Centrifuge clinical body fluid samples at 20,000 × g for 15 minutes.
- cfDNA Extraction: Extract cell-free DNA from 400 μL of supernatant using a specialized cfDNA kit (e.g., VAHTS Free-Circulating DNA Maxi Kit).
- wcDNA Extraction: Add lysis beads to the retained precipitate and shake vigorously to facilitate cell lysis. Extract whole-cell DNA from the precipitate using a standard DNA mini kit (e.g., Qiagen DNA Mini Kit).
- Library Preparation & Sequencing: Prepare DNA libraries for both cfDNA and wcDNA. Sequence on a high-throughput platform (e.g., Illumina NovaSeq) with at least 10 million reads per sample.
- Bioinformatic Analysis: Analyze data to determine the proportion of host vs. microbial reads and identify reportable pathogens using established criteria.
The table below summarizes key findings from recent studies comparing different methods.
| Method | Host DNA Proportion | Sensitivity / Concordance with Culture | Key Advantage |
|---|---|---|---|
| wcDNA mNGS | Mean: 84% [20] | 74.07% Sensitivity; 63.33% Concordance [20] | Higher sensitivity for pathogen detection [20]. |
| cfDNA mNGS | Mean: 95% [20] | 46.67% Concordance [20] | -- |
| 16S rRNA NGS | -- | 58.54% Concordance [20] | -- |
| ZISC-Filtered gDNA mNGS | >10x increase in microbial reads (9351 RPM vs. 925 RPM in unfiltered) [5] [19] | 100% detection in culture-positive sepsis samples (8/8) [5] [19] | Effectively enriches microbial content from blood. |
Workflow Visualization
The Scientist's Toolkit: Research Reagent Solutions
| Product / Technology | Function | Application in Host/Pathogen NGS |
|---|---|---|
| ZISC-based Filtration Device (e.g., Devin filter) | Pre-extraction physical removal of host white blood cells via a specialized coating. | Enriches microbial content in blood samples by depleting >99% of host cells, significantly reducing host DNA background [5] [19]. |
| VAHTS Free-Circulating DNA Maxi Kit | Extraction of cell-free DNA (cfDNA) from plasma or other liquid supernatants. | Used for preparing libraries for cfDNA-based mNGS, which can help detect pathogens but may have lower sensitivity compared to wcDNA approaches [20]. |
| Qiagen DNA Mini Kit | Extraction of high-quality whole-cell DNA from cell pellets or tissues. | Used for preparing libraries for wcDNA-based mNGS, which has been shown to have higher sensitivity for pathogen detection in body fluids [20]. |
| NEBNext Microbiome DNA Enrichment Kit | Post-extraction depletion of CpG-methylated host DNA. | An alternative method to reduce host DNA background by leveraging differences in methylation patterns between host and microbial DNA [19]. |
| Ultra-Low Library Prep Kit | Preparation of sequencing libraries from samples with low microbial biomass. | Essential for generating high-quality NGS libraries from samples where pathogen nucleic acid is scarce relative to host material [19]. |
| ZymoBIOMICS Reference Material | Defined microbial community standards spiked with known quantities of bacteria and fungi. | Serves as an internal spike-in control to monitor the efficacy of the host depletion workflow and the sensitivity of pathogen detection throughout the process [19]. |
Host Depletion Methodologies: From Laboratory Techniques to Bioinformatics
FAQs and Troubleshooting Guides
General Principles
Q: Why are physical separation methods like filtration and centrifugation critical in chemogenomic NGS? In samples derived from a host (e.g., human tissues or blood), the vast majority of extracted nucleic acids are of host origin. This host DNA background can overwhelm sequencing capacity, drastically reducing the number of microbial reads and compromising the detection sensitivity for pathogens or other non-host organisms. Physical separation methods target the enrichment of microbial cells or DNA prior to sequencing [23].
Q: What is the fundamental difference between pre-extraction and post-extraction host DNA depletion? Pre-extraction methods physically separate microbial cells from host cells or degrade host DNA before the DNA extraction step. Examples include saponin lysis of human cells or nuclease digestion of free-floating host DNA. In contrast, post-extraction methods, such as enzymatic methylation-based depletion, selectively remove host DNA after total DNA (host and microbe) has been extracted [23].
Centrifugation
Q: My centrifuge is vibrating excessively during a run. What should I do? An unbalanced load is the most common cause of centrifuge vibration. Immediately turn off the centrifuge and ensure all sample tubes are of similar weight and are positioned opposite each other in the rotor. Also, inspect the rotor and centrifuge for any visible damage [24].
Q: Can I shorten centrifugation times to improve my workflow's turn-around-time? Yes, but this must be validated for your specific protocol. Some studies on clinical chemistry samples have found that reducing centrifugation time from 15 minutes to 7-10 minutes did not significantly alter test results, but this is highly dependent on the sample type and the relative centrifugation force (RCF) applied. Always refer to your specific protocol's requirements and validate any changes [25].
Q: The lid on my centrifuge won't lock. What could be wrong? Check for any physical obstructions preventing closure. Ensure the safety interlocks are functioning and inspect the lid gasket for tears or damage. If the gasket is damaged, do not use the centrifuge. Cleaning and lubricating the locking mechanism as per the manufacturer's manual may also help [24].
Filtration
Q: How does filtration work as a host depletion method? The F_ase method, for example, uses a 10 μm filter. This pore size allows smaller microbial cells to pass through or be captured while retaining larger mammalian host cells. The filtrate, enriched in microbial cells, is then subjected to nuclease digestion to degrade any remaining cell-free host DNA before microbial DNA extraction [23].
Q: What are the trade-offs of using filtration for host DNA depletion? While effective at increasing microbial read counts, filtration may underrepresent microbial species that are larger than the filter's pore size or those that tend to form clumps. It can also be less effective on samples with a high viscosity that may clog the filter [23].
Common Issues and Solutions
Q: I am consistently getting low yields after host DNA depletion and library preparation. What are the potential causes? Low yield can stem from multiple points in the workflow. The table below outlines common causes and corrective actions [21].
| Cause | Mechanism of Yield Loss | Corrective Action |
|---|---|---|
| Poor Input Quality | Sample contaminants inhibit enzymatic reactions. | Re-purify input sample; check absorbance ratios (260/280 ~1.8). |
| Overly Aggressive Cleanup | Desired DNA fragments are accidentally removed during bead-based cleanup. | Optimize bead-to-sample ratio; avoid over-drying beads. |
| Inefficient Ligation | Adapters do not ligate properly to insert DNA. | Titrate adapter-to-insert molar ratio; ensure fresh ligase. |
| Suboptimal Centrifugation | Incomplete pelleting or unwanted loss of material. | Balance loads properly; follow recommended RCF and time. |
Q: After centrifugation, my sample appears turbid. What does this indicate? In tissue lysates, turbidity often indicates the presence of indigestible protein fibers. These fibers can clog silica membranes during subsequent DNA purification, leading to low yield and protein contamination. The solution is to centrifuge the lysate at maximum speed for 3 minutes to pellet these fibers before proceeding with the binding steps [26].
Experimental Protocols for Host DNA Depletion
Protocol 1: Saponin Lysis and Nuclease Digestion (S_ase)
This pre-extraction method uses saponin to lyse host cells, followed by nuclease to degrade the released host DNA [23].
- Sample Preparation: Resuspend the sample in a buffer containing 0.025% saponin.
- Host Cell Lysis: Incubate the mixture to allow saponin to selectively lyse mammalian cells.
- Nuclease Digestion: Add a nuclease enzyme (e.g., Benzonase) to digest the liberated host DNA. Incubate for the recommended time.
- Nuclease Inactivation: Add EDTA to chelate cations and inactivate the nuclease.
- Microbial Pellet Recovery: Centrifuge the sample to pellet the intact microbial cells.
- Wash and Resuspend: Wash the pellet to remove lysis and digestion remnants, then resuspend in a suitable buffer for standard DNA extraction.
Protocol 2: Filtration and Nuclease Digestion (F_ase)
This pre-extraction method physically separates microbial cells from host cells using a filter [23].
- Sample Preparation: Dilute the sample if necessary to reduce viscosity.
- Filtration: Pass the sample through a 10 μm filter unit. Microbial cells pass through the filter or are captured on it, while larger host cells are retained.
- Filter Wash: Wash the filter with a buffer to recover any microbial cells.
- Nuclease Digestion: The flow-through and wash are combined, and a nuclease is added to digest any residual cell-free host DNA.
- Microbial DNA Extraction: Proceed with standard microbial DNA extraction from the nuclease-treated filtrate.
Performance Comparison of Host DNA Depletion Methods
The following table summarizes the performance of various host depletion methods as reported in a benchmark study on respiratory samples [23].
| Method | Type | Key Principle | Host DNA Load Post-Treatment (BALF) | Microbial Read Increase (BALF, fold) | Key Advantages/Disadvantages |
|---|---|---|---|---|---|
| S_ase | Pre-extraction | Saponin lysis + Nuclease | 493.82 pg/mL (0.011‰ of original) | 55.8x | High host removal. Potential taxonomic bias. |
| K_zym | Pre-extraction | Commercial Kit (HostZERO) | 396.60 pg/mL (0.009‰ of original) | 100.3x | Most effective at increasing microbial reads. |
| F_ase | Pre-extraction | 10μm Filtration + Nuclease | Data not specified | 65.6x | Balanced performance. May lose large microbes. |
| R_ase | Pre-extraction | Nuclease Digestion | Data not specified | 16.2x | High bacterial DNA retention. Lower host removal. |
| O_pma | Pre-extraction | Osmotic Lysis + PMA | Data not specified | 2.5x | Least effective in increasing microbial reads. |
Centrifugation Condition Effects
A study on clinical samples showed that centrifugation time could be optimized without affecting analytical results [25].
| Centrifugation Condition | Relative Centrifugal Force (RCF) | Centrifugation Time | Impact on Test Results |
|---|---|---|---|
| Condition 1 | 2180 g | 15 min | Reference standard (WHO guideline) |
| Condition 2 | 2180 g | 10 min | No significant difference from 15 min |
| Condition 3 | 1870 g | 7 min | No significant difference from 15 min |
Workflow and Pathway Visualizations
Pre-extraction Host DNA Depletion Process
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in Host DNA Depletion |
|---|---|
| Saponin | A detergent that selectively lyses mammalian cell membranes by complexing with cholesterol, releasing host cellular contents while leaving many microbial cells intact [23]. |
| Nuclease Enzyme (e.g., Benzonase) | An endonuclease that digests all forms of DNA and RNA (linear, circular, single- and double-stranded). Used to degrade host DNA after lysis, leaving microbial DNA protected within intact cells [23]. |
| Propidium Monoazide (PMA) | A DNA-intercalating dye that penetrates only membrane-compromised (dead) cells. Upon photoactivation, it cross-links DNA, rendering it unamplifiable. Used in methods like O_pma to selectively remove DNA from lysed host cells [23]. |
| Silica Spin Columns | Used in DNA purification kits to bind DNA after host depletion steps. The silica membrane selectively binds DNA in the presence of high-salt buffers, allowing contaminants to be washed away [26]. |
| Magnetic Beads | Used for high-throughput DNA cleanup and size selection. The bead-to-sample ratio is critical for efficient recovery of the target DNA fragment size and removal of adapter dimers [21]. |
In chemogenomic next-generation sequencing (NGS) research, the presence of high levels of host DNA in samples from tissues, blood, or respiratory fluids presents a significant analytical challenge. Selective host DNA degradation through enzymatic and chemical methods enables researchers to deplete this background interference, thereby enriching microbial or pathogen DNA for more effective sequencing and analysis. This technical support center provides essential guidance for implementing these critical techniques.
Core Concepts and Methodologies
What is selective host DNA degradation and why is it critical for NGS?
Selective host DNA degradation refers to laboratory methods that preferentially remove or deplete DNA from the host organism (e.g., human DNA from a clinical sample) to improve the detection and analysis of non-host DNA, such as from pathogens or microbes [2]. This is a crucial sample preparation step for metagenomic NGS (mNGS) in clinical and research settings.
The necessity for this step arises because many clinical samples, like respiratory fluids, blood, or tissues, contain an overwhelming amount of host DNA. For instance, untreated bronchoalveolar lavage (BAL) and sputum samples can consist of 99.7% and 99.2% host reads, respectively [2]. Sequencing without host depletion results in a shallow effective sequencing depth for microbial DNA, severely underestimating microbial diversity and potentially missing critical pathogens [2].
How do enzymatic and chemical methods compare?
Different methods operate on distinct principles to achieve host DNA depletion. The table below summarizes the core mechanisms of common approaches:
| Method Type | Example | Core Mechanism |
|---|---|---|
| Enzymatic Digestion | Restriction Enzyme Digestion [27] | Uses restriction enzymes (e.g., BamHI, XmaI) to cut host DNA at specific sequence sites not present in the target parasite or microbial DNA, reducing host template amplification. |
| Enzymatic Depletion | Benzonase-based method [2] | Utilizes enzymes to degrade host DNA while protecting microbial DNA, often by exploiting differences in cell wall structures. |
| Commercial Kits (Multi-mechanism) | MolYsis, HostZERO, QIAamp [2] | Often employ a combination of enzymatic, chemical, and/or physical lysis steps to selectively lyse human cells and degrade the released DNA. |
| Physical Separation | ZISC Filtration [5] | A novel filtration device that physically depletes host white blood cells (WBCs) while allowing microbes to pass through for subsequent DNA extraction. |
Troubleshooting Guides & FAQs
Our lab is new to host depletion. What is the most common pitfall?
The most common pitfall is applying a single method universally across all sample types without optimization. The optimal host DNA depletion method is highly dependent on your sample type, the clinical question, and the target pathogens [28]. For example, a method optimized for frozen respiratory samples may not perform well for blood samples. It is crucial to optimize a specific workflow for each sample type and question you aim to address [28].
After host depletion and mNGS, our microbial reads are still very low. What could be wrong?
Low microbial reads post-depletion can stem from several issues in the workflow. Consider the following troubleshooting checklist:
- Inefficient Depletion Method: The chosen method may not be effective for your specific sample matrix. For example, some commercial kits like MolYsis and HostZERO showed variable efficiency across BAL, nasal, and sputum samples [2].
- Sample Input Quality: The initial microbial load may be too low, or the sample storage conditions may have reduced microbial viability. Freezing without cryoprotectants can reduce the viability of certain bacteria like Pseudomonas aeruginosa [2].
- Inhibition in Downstream Steps: Components from the host depletion kit may carry over into the DNA extraction or library preparation steps, inhibiting enzymatic reactions [29].
- Over-fragmentation of DNA: During library prep, over-digestion during enzymatic fragmentation can make DNA molecules too short for sequencing. Always follow protocol recommendations for incubation times [30].
We see a shift in the microbial community composition after host depletion. Is this a bias?
A shift in composition can occur and may represent both a true enrichment and a potential methodological bias. Host depletion increases the effective sequencing depth, revealing microbial species that were previously masked by host reads [2]. However, some methods can also introduce bias. For instance, one study noted that most methods did not change the community structure of BAL and nasal samples, but the proportion of Gram-negative bacteria decreased in sputum samples from people with cystic fibrosis after treatment [2]. Furthermore, enzymatic methods can sometimes exhibit sequence bias during fragmentation [30]. Always include appropriate controls to help distinguish true signal from bias.
How critical are controls in a host depletion workflow?
Controls are critical at every stage to ensure results are reliable and interpretable, especially given the high variability of clinical samples [28]. The table below outlines essential controls:
| Stage | Control Type | Purpose |
|---|---|---|
| Sample Collection | Negative Control (e.g., sterile swab, water) | Detect contamination introduced during sample taking or from the collection medium [28]. |
| DNA Extraction | Positive Control (External Quality Assurance sample) | Verify the method yields expected results and is reproducible across runs [28]. |
| Library Preparation | Negative Control (Reagent-only control) | Identify background contamination present in extraction or library prep kits (the "kitome") [28]. |
| Sequencing | Positive Control (Known mock community) | Confirm the entire wet-lab and bioinformatics pipeline is functioning correctly [28]. |
| Bioinformatics | In-silico Negative Control | Establish a baseline for background "noise" in the final data output [28]. |
Experimental Protocols & Data
Quantitative Comparison of Host Depletion Methods
The following table summarizes a head-to-head comparison of five host DNA depletion methods performed on frozen human respiratory samples, as reported in a 2024 study [2]. This data can guide your method selection.
| Method | Reduction in Host DNA (by Sample Type) | Increase in Final Microbial Reads (vs. Untreated) | Impact on Species Richness |
|---|---|---|---|
| lyPMA | Not the most effective for tested frozen samples [2]. | Not significant for BAL; increased for other types [2]. | Increased for some sample types [2]. |
| Benzonase | Less effective for nasal swabs [2]. | Increased for sputum [2]. | Increased for some sample types [2]. |
| MolYsis | ~69.6% decrease in sputum [2]. | ~100-fold increase in sputum; 10-fold in BAL [2]. | Significantly increased for BAL and nasal [2]. |
| HostZERO | ~73.6% decrease in nasal; ~45.5% in sputum [2]. | ~50-fold increase in sputum; 8-fold in nasal [2]. | Significantly increased for nasal [2]. |
| QIAamp | ~75.4% decrease in nasal [2]. | ~25-fold increase in sputum; 13-fold in nasal [2]. | Significantly increased for nasal [2]. |
Detailed Protocol: Restriction Enzyme-Based Host DNA Depletion
This protocol is adapted from a method validated for detecting blood-borne parasites via 18S rRNA gene sequencing [27].
- Principle: Restriction enzymes (BamHI and XmaI) are used to digest the host 18S rRNA gene template at cut sites present in the host sequence but absent in the target parasite sequences. This reduces host template competition during subsequent PCR amplification [27].
- Workflow:
- DNA Extraction: Extract total DNA from the sample (e.g., blood) using a standard kit (e.g., Qiagen Blood Mini Kit).
- Restriction Digestion:
- Prepare a digestion reaction with the extracted DNA and the restriction enzymes (e.g., BamHI and XmaI).
- Incubate according to the enzymes' optimal conditions (temperature and time).
- DNA Clean-up: Post-digestion, clean the DNA using a PCR clean-up kit (e.g., Monarch PCR & DNA Cleanup Kit). The protocol in [27] recommends splitting the cleaned DNA to select for both >2 kb and <2 kb products to account for varying parasite DNA fragment sizes.
- PCR Amplification: Amplify the target gene region (e.g., ~200 bp of the 18S rRNA gene) using universal primers.
- Library Prep and Sequencing: Proceed with standard NGS library preparation and sequencing.
- Key Validation Point: This method led to a substantial reduction in human reads and a corresponding 5- to 10-fold increase in parasite reads relative to undigested samples, allowing for discrimination of mixed parasitic infections [27].
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Kit Name | Function in Host DNA Depletion | Applicable Sample Types |
|---|---|---|
| MolYsis Kits (e.g., Basic5, Complete5) [28] | Selective lysis of human cells and degradation of released DNA; some kits integrate microbial DNA extraction. | Liquid samples (e.g., blood, BAL) [28]. |
| HostZERO Microbial DNA Kit [2] | Commercial kit for depleting host DNA to improve microbial sequencing. | Respiratory samples (e.g., nasal, sputum, BAL) [2]. |
| QIAamp DNA Microbiome Kit [2] | Commercial kit that depletes host DNA while enriching microbial DNA. | Respiratory samples; shown to minimally impact Gram-negative bacteria viability [2]. |
| Benzonase-based Method [2] | An enzymatic approach tailored for degrading host DNA in specific matrices like sputum. | Sputum, skin swabs, saliva [2]. |
| Restriction Enzymes (BamHI, XmaI) [27] | Digests host DNA at specific sequence sites to reduce template competition in PCR-based NGS. | Blood samples for parasite detection [27]. |
| Devin Filter (ZISC) [5] | A novel filtration device that physically removes host white blood cells (>99%) while preserving microbes. | Blood samples for sepsis diagnostics [5]. |
Workflow Visualization
Diagram Title: Host DNA Depletion Method Selection Workflow
Zwitterionic Interface Ultra-Self-assemble Coating (ZISC) technology represents a significant advancement in biomedical filtration and coating methods. Inspired by the surface arrangement of the cell-lipid bilayer, zwitterionic materials create a protective layer on material surfaces that prevents contact with biological substances while maintaining strong hydrophilicity and high biocompatibility [31]. The technology is characterized by its high hydrophilicity, low surface free energy, strong hydration, and weak biomolecule interactions, resulting in adhesion resistance to common biological substances [31].
For researchers in chemogenomic next-generation sequencing (NGS), the primary application of ZISC technology lies in its ability to efficiently deplete host cells from biological samples, thereby significantly reducing human DNA background and improving microbial signal detection in metagenomic NGS (mNGS) [1]. This addresses a critical challenge in clinical diagnostics where the overwhelming abundance of human DNA consumes valuable sequencing capacity and masks pathogenic signals.
Technical FAQs and Troubleshooting
Q: What filtration efficiency can I expect from ZISC-based filters for white blood cell removal? A: ZISC-based filters consistently achieve >99% white blood cell (WBC) removal across various blood volumes while allowing unimpeded passage of bacteria and viruses [1]. This high efficiency is maintained across different blood volumes (3-13 mL in validation studies) and is crucial for effective host DNA depletion in mNGS workflows.
Q: How does ZISC-based filtration compare to other host depletion methods? A: Research demonstrates ZISC-based filtration outperforms alternative host depletion techniques in both efficiency and practicality:
Table: Comparison of Host Depletion Methods
| Method | Mechanism | Efficiency | Practical Considerations |
|---|---|---|---|
| ZISC-based Filtration | Physical filtration with zwitterionic-cell binding | >99% WBC removal [1] | Less labor-intensive, preserves microbial integrity [1] |
| Differential Lysis (QIAamp Kit) | Chemical lysis of human cells | Variable efficiency | Complex workflow, may damage some microbes [1] |
| CpG-Methylated DNA Removal (NEBNext Kit) | Enzymatic removal of methylated host DNA | Post-extraction only | Doesn't prevent host DNA from consuming extraction resources [1] |
Q: Why is my post-filtration microbial recovery inconsistent? A: Inconsistent recovery typically stems from two main issues:
- Filter clogging: Ensure you're using the patented ZISC coating which prevents clogging regardless of filter pore size [1]
- Sample processing speed: Maintain gentle plunger depression when using syringe-based filtration; aggressive pressure can damage microbial cells [1]
Q: What performance improvement should I expect in my mNGS workflow? A: Clinical validations demonstrate substantial improvements:
- gDNA-based mNGS with ZISC filtration detected all expected pathogens in 100% (8/8) of clinical samples [1]
- Average microbial read count of 9,351 reads per million (RPM), over tenfold higher than unfiltered samples (925 RPM) [1]
- Microbial composition remains unaltered, ensuring accurate pathogen profiling [1]
Q: Can ZISC technology be applied to different sample types beyond blood? A: While most extensively validated for blood samples, the fundamental principles of zwitterionic interaction with biological components suggest potential application to various sample types. However, optimal performance requires validation with your specific sample matrix as binding efficiencies may vary.
Experimental Protocols and Workflows
Standard ZISC Filtration Protocol for Blood Samples
Materials Required:
- ZISC-based fractionation filter (commercially available as Devin filter from Micronbrane)
- Syringe (appropriate for sample volume)
- Collection tube (15 mL Falcon tube recommended)
- Low-speed centrifuge
- High-speed centrifuge (capable of 16,000g)
- ZISC-based Microbial DNA Enrichment Kit or equivalent
Procedure:
- Transfer approximately 4 mL of whole blood to a syringe
- Securely connect the ZISC-based filter to the syringe
- Gently depress the syringe plunger, pushing blood sample through filter into collection tube
- Subject filtered blood to low-speed centrifugation (400g for 15 min at room temperature) to isolate plasma
- Transfer plasma to new tube for high-speed centrifugation (16,000g) to obtain microbial pellet
- Extract DNA using appropriate kit (ZISC-based Microbial DNA Enrichment Kit recommended)
- Proceed with standard mNGS library preparation
Validation Protocol for Filter Performance
Purpose: Verify filter efficiency and microbial recovery Materials:
- Complete blood cell count analyzer
- Standard plate-enumeration materials
- qPCR setup for viral quantification
- Spiked controls (E. coli, S. aureus, K. pneumoniae recommended)
Procedure:
- Pre-filtration: Measure WBC count using blood analyzer
- Process blood sample through ZISC filter
- Post-filtration: Measure WBC count in filtrate
- Calculate depletion efficiency: [(Pre-count - Post-count)/Pre-count] × 100%
- For microbial passage: Spike blood with 10⁴ CFU/mL of control organisms
- Filter and quantify bacterial counts in filtrate using plate enumeration
- For viral passage: Spike with feline coronavirus and quantify using qPCR
Research Reagent Solutions
Table: Essential Materials for ZISC-Based Host Depletion Workflows
| Reagent/Material | Function | Application Notes |
|---|---|---|
| ZISC-based Fractionation Filter | Host cell depletion | >99% WBC removal; preserves microbial integrity [1] |
| ZISC-based Microbial DNA Enrichment Kit | DNA extraction from filtered samples | Optimized for post-filtration processing [1] |
| ZymoBIOMICS Reference Materials (D6320, D6331) | Process controls and spike-in controls | Validate microbial recovery; D6331 contains 21 bacterial/fungal species [1] |
| Ultra-Low Library Prep Kit | mNGS library preparation | Compatible with low-biomass samples post-filtration [1] |
Performance Data and Metrics
Table: Quantitative Performance of ZISC-based Filtration in mNGS
| Parameter | Unfiltered Samples | ZISC-Filtered Samples | Improvement Factor |
|---|---|---|---|
| Microbial RPM | 925 RPM [1] | 9,351 RPM [1] | >10-fold |
| Pathogen Detection Rate | Variable, culture-dependent | 100% (8/8 clinical samples) [1] | Significant enhancement |
| WBC Depletion | Baseline | >99% [1] | Essential for host DNA reduction |
| Genome Coverage | Limited by host background | Up to 98.9% achievable [7] | Dependent on initial pathogen load |
Advanced Applications and Future Directions
Recent studies have expanded ZISC technology applications beyond sepsis diagnosis. In pulmonary tuberculosis diagnosis, host DNA depletion-assisted mNGS (HDA-mNGS) demonstrated significantly improved detection sensitivity (72.0% vs 51.2% with conventional mNGS) in bronchoalveolar lavage fluid samples [16]. The technology also provided increased coverage of the MTB genome by up to 16-fold and enhanced detection of antimicrobial resistance loci [16].
For SARS-CoV-2 detection, host DNA-removed mNGS achieved 92.9% detection rate in samples with Ct value ≤35 while simultaneously enabling analysis of host local immune signaling [7]. This dual capability of comprehensive pathogen identification and host response analysis represents a significant advantage for research applications.
The fundamental mechanism involves zwitterionic polymers creating a highly hydrated interface through electrostatic and hydrogen bonding with water molecules, forming a protective layer that resists protein adsorption and cell adhesion [31]. The specific capture of white blood cells is achieved through careful design of charge bias on the zwitterionic surface, creating selective affinity while allowing other blood components to pass through unimpeded [31].
Bioinformatics filtering is a critical post-sequencing step for reducing host DNA background in chemogenomic Next-Generation Sequencing (NGS) research. When physical or enzymatic host depletion methods are applied during sample preparation, a significant proportion of host sequences often remains in the sequencing data, particularly in low-biomass samples or those with extremely high initial host content, such as blood and tissue [32] [33]. Computational methods provide a final, vital defense by identifying and removing these residual host reads, thereby enriching the dataset for microbial or pathogenic signals and significantly improving the sensitivity of downstream analyses [33]. This guide details the methodologies, tools, and best practices for implementing effective bioinformatics host sequence removal.
Key Bioinformatics Tools and Workflows
The core task of bioinformatics host filtering involves aligning sequencing reads to a reference host genome and discarding those that map to it. The following table summarizes the primary tools and their key characteristics.
Table 1: Key Bioinformatics Tools for Host Sequence Removal
| Tool Name | Primary Function | Key Features | Applicable Data Types |
|---|---|---|---|
| KneadData [33] | Integrated filtering pipeline | Combines quality trimming (Trimmomatic) and host read removal (Bowtie2). Includes pre-built databases for human and mouse genomes. | Short-read (Illumina) |
| Bowtie2 [33] | Read alignment | A fast and memory-efficient tool for aligning sequencing reads to large reference genomes, such as the human genome. | Short-read (Illumina) |
| BWA (Burrows-Wheeler Aligner) [33] | Read alignment | A highly accurate alignment tool, particularly suitable for high-throughput sequencing data for host read subtraction. | Short-read (Illumina) |
| BMTagger [33] | Human sequence removal | A tool developed by NCBI specifically for detecting and tagging sequences originating from human contamination in microbiome data. | FASTA, FASTQ, SRA |
| CLEAN [34] | All-in-one decontamination | Removes host sequences, spike-in controls (e.g., PhiX), and rRNA. Works with both short- and long-read technologies (Illumina, Nanopore). | Short-read, Long-read, FASTA |
The general workflow for host sequence removal follows a logical pipeline from raw sequencing data to cleaned data ready for microbial analysis.
Experimental Protocols and Performance Benchmarks
Protocol: Standard Host Read Removal Using KneadData and Bowtie2
This protocol is commonly used for processing short-read metagenomic data [33].
- Input: Raw sequencing reads in FASTQ format (single-end or paired-end).
- Quality Trimming: Use Trimmomatic (integrated within KneadData) to remove adapter sequences and low-quality bases from the reads. Example parameters include a minimum read length of 50 bp and a minimum quality score of 20.
- Host Alignment: Use Bowtie2 to align the trimmed reads against a host reference genome (e.g., human GRCh38). The alignment process is optimized for speed and sensitivity.
- Read Separation: The output of the alignment is processed to separate reads that map to the host genome (to be discarded) from those that do not map (non-host reads).
- Output: The final output is a set of cleaned FASTQ files containing the non-host reads, which are enriched for microbial sequences and suitable for taxonomic profiling or functional analysis.
Quantitative Data on Host Depletion Impact
Empirical studies demonstrate the profound impact of combined wet-lab and computational host depletion. The following table summarizes key performance metrics from recent research.
Table 2: Impact of Host DNA Removal on Metagenomic Analysis
| Study & Sample Type | Method | Key Metric | Result with Host DNA Removal | Control (No Removal) |
|---|---|---|---|---|
| Sepsis Blood Samples [5] | Novel Filtration (gDNA mNGS) + Bioinformatics | Microbial Read Count (RPM) | ~9,351 RPM | ~925 RPM |
| Human/Mouse Colon Biopsies [33] | Host DNA Removal + Bioinformatics | Bacterial Species Detected per Sample | Significantly Increased | Baseline (Lower) |
| Human/Mouse Colon Biopsies [33] | Host DNA Removal + Bioinformatics | Bacterial Gene Detection Rate | Increased by 33.89% (Human) & 95.75% (Mouse) | Baseline |
Troubleshooting Common Issues
Problem: Incomplete Host Read Removal After Filtering
- Potential Cause: The host reference genome used is incomplete or does not match the host sample (e.g., using a standard human reference for a sample with known significant genetic variations) [33].
- Solution: Ensure you are using the most comprehensive and appropriate reference genome available. For human samples, use the latest build from the Genome Reference Consortium.
Problem: Low Microbial Read Recovery After Host Filtering
- Potential Cause: Overly stringent alignment parameters during the host filtering step can cause microbial reads with slight homology to the host genome to be incorrectly discarded [32] [34].
- Solution: Use standardized parameters from established pipelines like KneadData. For advanced users, consider tuning alignment stringency (e.g.,
--very-sensitivein Bowtie2) and validating results with a mock microbial community.
Problem: Persistent Contamination from Reagents or Spike-ins
- Potential Cause: Standard host filtering tools are designed to remove host sequences but may not target common laboratory contaminants or intentionally added control sequences (e.g., PhiX, DCS amplicon) [34] [35].
- Solution: Use a comprehensive decontamination pipeline like CLEAN, which allows for the simultaneous removal of host sequences and common spike-in contaminants using a combined reference file [34].
Problem: Challenges with Long-Read Sequencing Data
- Potential Cause: Traditional alignment tools like Bowtie2 and BWA are optimized for short reads and may not handle long-read data (Oxford Nanopore, PacBio) effectively.
- Solution: Employ tools specifically designed for long-read data. The CLEAN pipeline, for instance, uses
minimap2for alignment, which is suitable for both long and short reads [34].
Frequently Asked Questions (FAQs)
Q1: Can bioinformatics filtering completely replace experimental host DNA depletion methods? No, it is most effective as a complementary step. Experimental methods (e.g., filtration, enzymatic digestion) reduce the host DNA burden upfront, making sequencing more cost-effective by preventing the allocation of a large majority of reads to host DNA. Bioinformatics filtering then serves as a final, precise cleaning step to remove any residual host sequences [33]. Relying solely on bioinformatics filtering after sequencing a sample with >99% host DNA is computationally wasteful and may fail to detect very low-abundance microbes.
Q2: What are the primary limitations of bioinformatics host filtering? The two main limitations are:
- Dependence on Reference Genomes: The method can only remove sequences that are present in the provided host reference genome. It cannot remove host sequences that are novel or highly divergent from the reference [33].
- Inability to Remove Homologous Sequences: Reads from microbial genes that share homology with host genes may be incorrectly identified as host and removed, potentially leading to the loss of biologically relevant signals [33].
Q3: How can I identify and manage contamination from laboratory reagents or cross-sample contamination in my data? Contamination is a significant challenge in low-biomass studies [36]. Key strategies include:
- Using Negative Controls: Process negative controls (e.g., blank water extracts) alongside your samples through the entire workflow, from extraction to sequencing [36] [35].
- Bioinformatic Identification: Tools like
Decontam(for R) use prevalence or frequency-based statistical methods to identify contaminants by comparing their abundance in true samples versus negative controls [34] [35]. - Standardized Reporting: Adhere to emerging guidelines for reporting contamination and removal workflows in microbiome studies to ensure reproducibility [36].
Q4: We are working with RNA-Seq data from host cells. Is this workflow relevant? Yes, the principle is similar. For host RNA-Seq data, a common goal is to remove ribosomal RNA (rRNA) reads to improve the resolution of mRNA sequencing. Pipelines like CLEAN can be configured to map reads to an rRNA reference database and remove those that align, leaving behind enriched mRNA sequences for downstream expression analysis [34].
Table 3: Key Resources for Bioinformatics Host Depletion
| Item | Function in Host Depletion | Example/Note |
|---|---|---|
| Host Reference Genome | The sequence against which reads are aligned to identify and remove host-derived data. | Human: GRCh38 (hg38); Mouse: GRCm39 (mm39). |
| KneadData Pipeline | An integrated, user-friendly pipeline that performs both quality control and host read removal. | Includes built-in host databases; good for users seeking a standardized workflow [33]. |
| CLEAN Pipeline | A comprehensive, reproducible pipeline for removing host sequences, spike-ins, and rRNA from various data types. | Ideal for complex decontamination needs and long-read data [34]. |
| Negative Control Data | Sequencing data from blank extractions used to identify contaminating sequences present in reagents. | Essential for reliable interpretation of low-biomass microbiome data [36] [35]. |
| High-Performance Computing (HPC) Cluster | Provides the computational power needed for aligning millions of reads against large reference genomes. | Necessary for processing large datasets in a timely manner. |
FAQs and Troubleshooting Guides
How can I overcome excessive human DNA background in blood samples for mNGS?
Excessive host DNA in blood samples is a major obstacle, but several host depletion methods can significantly improve microbial detection.
- Pre-extraction Filtration: Novel filtration technologies, such as the Zwitterionic Interface Ultra-Self-assemble Coating (ZISC)-based filter, can physically remove host white blood cells before DNA extraction. One study demonstrated that this method achieved >99% removal of white blood cells while allowing bacteria and viruses to pass through unimpeded. When applied to clinical sepsis samples, this resulted in a tenfold increase in microbial read counts compared to unfiltered samples [19] [5].
- Post-extraction Methylation-Based Enrichment: Commercial kits like the NEBNext Microbiome DNA Enrichment Kit and the MethylMiner Methylated DNA Enrichment Kit exploit the differential methylation patterns between host and microbial DNA. These kits bind and remove methylated host DNA, enriching for non-methylated microbial DNA. A comparative study showed that the NEBNext kit led to a significant decrease in human genome reads and a concurrent significant increase in reads from spiked-in bacteria like K. pneumoniae and S. aureus [37].
- Choosing the Right DNA Source: For blood samples, using genomic DNA (gDNA) from a cell pellet, rather than cell-free DNA (cfDNA) from plasma, is crucial for enabling effective pre-extraction host depletion. Research has shown that host depletion methods significantly enhance gDNA-based mNGS but provide minimal benefit for cfDNA-based approaches [19].
Troubleshooting Tip: If your blood mNGS results show low microbial read counts despite high sequencing depth, consider integrating a pre-extraction host depletion step. The ZISC-based filtration method is noted for being less labor-intensive than some alternative methods [19].
What are the best practices for respiratory sample collection and analysis to ensure reliable mNGS results?
The quality of respiratory samples directly impacts the reliability of mNGS results, making proper collection and quality control paramount.
- Sample Quality Assessment: For sputum samples, use the Bartlett grading system to assess quality. Only samples with a score of ≤1 (indicating ≤10 squamous epithelial cells and ≥25 leukocytes per low-power field) should be used for mNGS. This minimizes contamination from oropharyngeal flora and ensures the sample originates from the lower respiratory tract [38].
- Appropriate Sample Type: While sputum is commonly used, Bronchoalveolar Lavage Fluid (BALF) is often the preferred specimen type because it is collected directly from the site of infection, reducing the potential for upper respiratory tract contamination. Studies have successfully used BALF mNGS to identify a wide range of pathogens, including bacteria, viruses, and fungi, with a high positive clinical impact [39] [40].
- Clinical Interpretation is Key: A positive mNGS result does not always indicate an active infection; it may represent colonization. Therefore, clinicians must correlate sequencing results with the patient's clinical symptoms, imaging findings, and other laboratory tests. A study on pulmonary infections found that a clinician committee was critical for interpreting mNGS results and guiding appropriate patient management [40].
Troubleshooting Tip: If your mNGS results from a respiratory sample show a high diversity of oral commensal bacteria, re-evaluate the sample's quality score. The sample may have been contaminated during collection, and the results should be interpreted with caution [38].
What are common library preparation failures and how can they be diagnosed and fixed?
Library preparation is a critical step where errors can lead to sequencing failure. Common issues fall into several categories [21]:
| Problem Category | Typical Failure Signals | Common Root Causes & Corrective Actions |
|---|---|---|
| Sample Input & Quality | Low yield; smear on electropherogram; low complexity [21]. | Causes: Degraded DNA/RNA; contaminants (phenol, salts); inaccurate quantification [21].Fixes: Re-purify input; use fluorometric (Qubit) over UV quantification; check 260/230 and 260/280 ratios [21]. |
| Fragmentation & Ligation | Unexpected fragment size; high adapter-dimer peak [21]. | Causes: Over-/under-shearing; improper adapter-to-insert ratio [21].Fixes: Optimize fragmentation parameters; titrate adapter concentration [21]. |
| Amplification (PCR) | High duplicate rate; amplification bias [21]. | Causes: Too many PCR cycles; enzyme inhibitors [21].Fixes: Reduce cycle number; use clean, high-quality input DNA [21]. |
| Purification & Cleanup | High adapter-dimer signal; sample loss [21]. | Causes: Wrong bead-to-sample ratio; over-drying beads; pipetting error [21].Fixes: Precisely follow cleanup protocols; use master mixes to reduce pipetting errors [21]. |
Diagnostic Flow: To systematically diagnose a problem, (1) check the electropherogram for abnormal peaks (e.g., a sharp ~120 bp peak indicates adapter dimers), (2) cross-validate DNA quantification with both fluorometric and qPCR methods, and (3) trace the problem backward through each preparation step [21].
Experimental Protocols for Key Workflows
Protocol 1: ZISC-Based Host Depletion for Blood Samples
This protocol is adapted from a study optimizing mNGS for sepsis diagnosis [19].
Principle: A specialized filter coating selectively binds and retains host leukocytes based on their surface properties, allowing microbial cells to pass through for downstream processing.
Workflow Diagram:
Key Steps:
- Filtration: Transfer approximately 4 mL of fresh, anti-coagulated whole blood into a syringe attached to the ZISC-based filter. Gently depress the plunger to pass the blood through the filter into a collection tube [19].
- Plasma Separation: Centrifuge the filtered blood at 400g for 15 minutes at room temperature to separate the plasma from any remaining cells [19].
- Microbial Pellet Isolation: Transfer the plasma to a new tube and perform a high-speed centrifugation at 16,000g to pellet microbial cells and debris [19].
- DNA Extraction and mNGS: Proceed with DNA extraction from the pellet using a standard microbial DNA extraction kit. This gDNA is then used for library preparation and sequencing [19].
Expected Outcome: This protocol should achieve >99% depletion of white blood cells, leading to a dramatic reduction in host DNA background and a significant (over tenfold) enrichment of microbial reads in the final sequencing data [19] [5].
Protocol 2: Methylation-Based Host DNA Depletion for Various Samples
This protocol compares methods suitable for samples like respiratory fluids or tissue homogenates, based on a comparative study of host depletion methods [37].
Principle: Human DNA is rich in methylated cytosine bases (CpG methylation), while most microbial DNA is not. This difference is exploited to selectively remove host sequences.
Workflow Diagram:
Key Steps and Method Comparison:
| Depletion Method | Principle | Input DNA Requirement | Key Procedural Steps |
|---|---|---|---|
| NEBNext Microbiome DNA Enrichment Kit | MBD2 protein bound to magnetic beads captures methylated host DNA [37]. | High molecular weight (≥15 kb), non-fragmented [37]. | 1. Incubate DNA with MBD2-bound beads.2. Apply magnet.3. Recover supernatant containing enriched microbial DNA [37]. |
| MethylMiner Kit | MBD2 protein coupled to streptavidin beads captures methylated DNA [37]. | Fragmented DNA (<1000 bp) [37]. | 1. Fragment DNA.2. Incubate with MBD2-beads.3. The microbial DNA is in the wash-through fraction; host DNA is bound to beads [37]. |
| MspJI Restriction Enzyme | Enzyme digestion cuts methylated DNA for depletion [37]. | Fragmented or non-fragmented [37]. | 1. Digest DNA with MspJI.2. For non-fragmented DNA, run product on a gel and excise/purify the high molecular weight (undigested microbial) band [37]. |
Expected Outcome: The NEBNext kit has been shown to cause a significant decrease in human genome reads and a significant increase in bacterial reads. The MethylMiner kit can significantly improve the detection and genome coverage of certain fungi and bacteria [37].
The Scientist's Toolkit: Research Reagent Solutions
The following table details key reagents and kits used in the featured host depletion methods.
| Research Reagent / Kit | Primary Function | Key Features & Considerations |
|---|---|---|
| ZISC-based Filtration Device | Pre-extraction physical depletion of host white blood cells from whole blood [19]. | >99% WBC removal; preserves microbial integrity; less labor-intensive; compatible with gDNA-based mNGS [19]. |
| NEBNext Microbiome DNA Enrichment Kit | Post-extraction depletion of methylated host DNA [19] [37]. | Uses MBD2-Fc protein; requires intact, high molecular weight DNA; shown to significantly increase bacterial read counts [37]. |
| MethylMiner Methylated DNA Enrichment Kit | Post-extraction depletion of methylated host DNA from fragmented samples [37]. | Uses MBD2 protein on magnetic beads; requires fragmented DNA input; effective for enriching fungal and bacterial DNA [37]. |
| MspJI Restriction Endonuclease | Post-extraction digestion and depletion of methylated host DNA [37]. | Digests methylated CpG sites; requires post-digestion purification (e.g., gel extraction); can be used on fragmented or non-fragmented DNA [37]. |
| Agencourt AMPure XP Beads | Post-reaction purification and size selection for NGS libraries [37]. | Used for cleaning up and concentrating DNA after various steps (digestion, enrichment); critical for removing adapter dimers and selecting the correct fragment size [21] [37]. |
Core Principles: The Yield-Integrity Balance in NGS
The success of next-generation sequencing (NGS), particularly in applications like chemogenomics where distinguishing host from pathogen DNA is critical, hinges on the quality of the input genetic material. The fundamental challenge lies in balancing two often competing objectives: maximizing DNA yield and preserving DNA integrity. Achieving this balance is the cornerstone of reliable and sensitive sequencing data.
The presence of excessive host DNA poses a significant barrier to sensitive pathogen detection. In metagenomic NGS (mNGS) of blood samples, host DNA can constitute over 95% of the sequenced material, drastically reducing the reads available for identifying pathogenic organisms and thereby impairing diagnostic sensitivity [16] [19]. The impact of host DNA is so profound that it can necessitate extreme sequencing depths to achieve meaningful microbial coverage; one study noted that samples with 90% host DNA required substantially deeper sequencing to detect low-abundance species effectively [4].
Furthermore, the physical integrity of the DNA is paramount, especially for long-read sequencing technologies (e.g., PacBio, Oxford Nanopore). High-Molecular-Weight (HMW) DNA is defined by fragment lengths greater than 50 kilobases (kb), with optimal sizes often exceeding 100 kb. This integrity is crucial for accurate genome assembly, detection of structural variants, and navigating complex genomic regions [41]. However, HMW DNA is highly susceptible to fragmentation from mechanical forces, improper handling, and chemical degradation.
Host DNA Depletion Methodologies
Overcoming the host DNA background is a primary focus in optimizing NGS for infectious disease diagnostics and research. The following table summarizes the key host depletion strategies, their mechanisms, and performance characteristics.
Table 1: Comparison of Host DNA Depletion Methods for NGS
| Method | Working Principle | Reported Efficacy | Key Advantages | Key Limitations |
|---|---|---|---|---|
| Physical Filtration (ZISC-based) [19] | Selectively binds and retains host leukocytes based on surface chemistry, allowing microbes to pass through. | >99% removal of white blood cells; >10-fold increase in microbial reads. | High efficiency; preserves microbial composition; less labor-intensive. | Requires specialized filter device; may not retain intracellular pathogens. |
| Enzymatic Digestion (CpG Methylation-Based) [19] | Uses enzymes to selectively digest CpG-methylated host DNA. | Varies; can be effective but may not match filtration efficiency. | Post-extraction method; can be applied after DNA is isolated. | Risk of incomplete digestion; may not be cost-effective for many samples. |
| Differential Lysis [19] | Uses mild detergents to lyse human cells, followed by centrifugation to remove host DNA. | Lower efficiency compared to novel filtration methods. | Relatively simple protocol. | Can also lyse some pathogen types; risk of co-precipitating host and pathogen DNA. |
| Saponin-Based Host Depletion [16] | Treatment with saponin to lyse mammalian cells without damaging bacterial cell walls. | Significantly improved sensitivity for detecting Mycobacterium tuberculosis (72.0% vs 51.2% for conventional mNGS). | Effective for intracellular bacteria. | Protocol optimization required for different sample types. |
| Probe-Based Hybridization | Uses custom probes to bind and remove host DNA sequences. | Not directly evaluated in provided results. | High specificity for targeted host sequences. | High cost; complex protocol; requires prior knowledge of host genome. |
Optimized Experimental Protocols
Protocol 1: HMW DNA Extraction for Long-Read Sequencing
This protocol is optimized for obtaining long, intact DNA fragments essential for platforms like PacBio and Oxford Nanopore [41].
- Sample Preparation: Use fresh or flash-frozen tissues. For blood, collect in K2-EDTA tubes and process within days at 4°C or store long-term at -80°C. For tough plant tissues, use a pre-chilled mortar and pestle with liquid nitrogen to create a fine powder.
- Lysis: Employ gentle lysis buffers. For plants, a CTAB-based buffer (e.g., 4% CTAB, 1.4M NaCl, 0.1M Tris, 0.02M EDTA) supplemented with 1% β-mercaptoethanol is highly effective for neutralizing polysaccharides and polyphenols [42] [43]. Incubate at 65°C for 40 minutes to 2 hours.
- Purification: Use specialized HMW DNA kits (e.g., Nanobind magnetic disk kits) instead of conventional spin columns, which cause shearing. If using chloroform:isoamyl alcohol (24:1) purification, mix by gentle inversion, not vortexing.
- Precipitation and Washing: Precipitate DNA using isopropanol. Wash the pellet with 70% ethanol.
Elution: Gently resuspend the purified DNA in ultrapure water or elution buffer. Let it dissolve at room temperature for several hours or overnight at 4°C, avoiding pipetting.
Critical Handling Notes:
- Always use wide-bore pipette tips to minimize mechanical shearing.
- Avoid vortexing at any step after cell lysis. Mix by gentle inversion or tapping.
- Limit freeze-thaw cycles, as ice crystals fragment DNA. Store aliquots at consistent -80°C.
Protocol 2: Host DNA-Depleted mNGS from Whole Blood
This workflow utilizes a novel filtration device to deplete host cells prior to DNA extraction, dramatically improving pathogen detection [19].
- Sample Collection: Collect whole blood in appropriate anticoagulant tubes.
- Host Cell Depletion: Pass approximately 4 mL of whole blood through a ZISC-based filtration device (e.g., "Devin" filter from Micronbrane) using a syringe. The filter retains >99% of white blood cells while allowing bacteria and viruses to pass through unimpeded.
- Microbial Pellet Collection: Centrifuge the filtrate at low speed (400g for 15 min) to isolate plasma. Then, perform a high-speed centrifugation (16,000g) of the plasma to pellet the microbial cells.
- DNA Extraction: Extract DNA from the microbial pellet using a standard microbial DNA kit. The resulting DNA will be highly enriched for pathogen sequences.
- Library Preparation and Sequencing: Proceed with standard mNGS library prep and sequencing. The study validated this on both Illumina (MiSeq, NovaSeq) and Nanopore (MinION) platforms.
The following workflow diagram illustrates the key steps and decision points in the host-depleted mNGS protocol.
Troubleshooting FAQs
Q1: My DNA yield is high, but my sequencing libraries are failing. What could be wrong? This is a classic sign of poor DNA purity or integrity. High absorbance at A230 on a spectrophotometer indicates contamination with salts, solvents, or carbohydrates, which can inhibit enzymatic reactions in library prep [41] [43]. For metabolite-rich samples like plants, incorporate a sorbitol wash step into your CTAB protocol to remove these contaminants [43]. Furthermore, assess DNA integrity using a Fragment Analyzer or pulsed-field gel electrophoresis (PFGE); a low DNA Integrity Number (DIN) or smeared gel indicates fragmentation, which will lead to poor library efficiency.
Q2: I am working with sputum/BALF samples for TB diagnosis. My mNGS is not detecting Mycobacterium tuberculosis despite positive cultures. How can I improve detection? The extremely high host DNA background in these samples is likely masking the bacterial signal. Implement a host depletion step prior to DNA extraction. A saponin-based pre-treatment has been shown to significantly improve the sensitivity of mNGS for detecting intracellular M. tuberculosis in bronchoalveolar lavage fluid (BALF), increasing detection rates from 51.2% to 72.0% [16]. This method helps lyse human cells while preserving the integrity of the tough mycobacterial cell wall.
Q3: I need high-molecular-weight DNA for long-read sequencing, but my extracts are always fragmented. What are the most critical steps to check? The most common causes are mechanical shearing and inappropriate kits. First, eliminate all vortexing after lysis and use only wide-bore pipette tips for handling DNA [41]. Second, standard silica-column kits are not suitable for HMW DNA as they cause significant shearing; switch to a kit specifically designed for long-read sequencing, such as those using magnetic disk technology (e.g., Nanobind). Finally, ensure your starting tissue is freshly frozen and avoid repeated freeze-thaw cycles of the extracted DNA.
Q4: How does host DNA depletion actually improve mNGS results? Host DNA depletion improves mNGS in two key ways:
- Enrichment of Microbial Reads: By physically removing host cells or host DNA, a much larger proportion of the sequencing reads in your library will originate from microbes. One study reported an average of 9,351 microbial reads per million (RPM) in filtered blood samples versus only 925 RPM in unfiltered samples—a more than tenfold enrichment [19].
- Increased Genome Coverage: With more sequencing power directed at the pathogen, the coverage of its genome increases dramatically. Host depletion methods have been shown to increase coverage of the M. tuberculosis genome by up to 16-fold, which also enhances the ability to detect antibiotic resistance genes [16].
The Scientist's Toolkit: Essential Reagents & Kits
Table 2: Key Reagents and Kits for DNA Extraction Optimization
| Item / Kit Name | Type | Primary Function | Key Features / Applications |
|---|---|---|---|
| CTAB (Cetyltrimethylammonium bromide) [42] [43] | Chemical Reagent | Efficient lysis of plant and microbial cells; co-precipitation and removal of polysaccharides. | Ideal for challenging, metabolite-rich samples; often used with β-mercaptoethanol to inhibit oxidation. |
| β-mercaptoethanol (BME) [42] [43] | Antioxidant | Reduces disulfide bonds and inhibits polyphenolic oxidation, preventing DNA browning. | Critical component of CTAB buffer for plants and other polyphenol-rich tissues. |
| Mag-Bind Blood DNA HV Kit [44] | Commercial Kit | Automated or semi-automated isolation of genomic DNA from large-volume blood samples (up to 4 mL). | Optimized for biobanking applications; compatible with platforms like MagBinder Fit24. |
| Nanobind CBB / PanDNA Kits [41] | Commercial Kit | Gentle isolation of HMW DNA for long-read sequencing from blood, cells, and tissue. | Magnetic disk technology minimizes shearing; includes Short Read Eliminator (SRE) to enrich for long fragments. |
| ZISC-based Filtration Device [19] | Hardware/Device | Physical depletion of host white blood cells from whole blood samples prior to DNA extraction. | Enables >99% host cell removal for mNGS pathogen enrichment; simple syringe-operated workflow. |
| QIAsymphony SP (DSP DNA Midi Kit) [45] | Automated System | Magnetic bead-based automated nucleic acid extraction. | High-throughput 96-well format; shown to produce high gDNA yields from challenging sample types like PAXgene blood. |
Optimizing Host Depletion: Addressing Technical Challenges and Method Selection
Frequently Asked Questions
Q1: What are the primary metrics used to evaluate host depletion efficiency? The primary metrics for evaluating host depletion efficiency include the percentage of host DNA before and after depletion, the fold-increase in microbial reads, and the retention rate of bacterial DNA. These are typically measured using quantitative PCR (qPCR) and sequencing data analysis. A successful depletion significantly reduces the host DNA proportion while maintaining the integrity and relative abundance of the microbial community [46] [2] [47].
Q2: Why does my host-depleted sample still show low microbial read counts after sequencing? Low microbial reads post-depletion can result from several factors: excessive loss of microbial DNA during the physical removal steps, incomplete lysis of microbial cells with tough walls, or a high proportion of cell-free microbial DNA in the original sample which is removed along with host DNA in pre-extraction methods. Optimizing sample-specific protocols and including controls can help identify the specific issue [46] [2].
Q3: My microbial community profile seems biased after host depletion. Is this normal? Some host depletion methods can introduce taxonomic bias. Methods that involve filtration may under-represent larger microbes or fungi, while enzymatic treatments can disproportionately affect species with more fragile cell walls. It is crucial to validate the chosen method using a mock microbial community relevant to your sample type to identify and account for any systematic biases [46] [2].
Q4: How does sample type influence the choice of host depletion method? Sample type is a critical factor. Respiratory samples like BALF have very high host content and require highly efficient methods. Infected tissues may need mechanical homogenization as a first step. Blood samples require methods that effectively remove white blood cells. A method that works well for one sample type may be inefficient or introduce significant bias for another [2] [47] [1].
Q5: What are the essential quality controls for a host depletion experiment? Essential quality controls include:
- Negative Controls: Process blank samples (e.g., saline, deionized water) through the entire workflow to monitor reagent and environmental contamination [46] [16].
- Positive Controls (Mock Communities): Use a defined mix of microbial species to assess bias, DNA loss, and contamination introduced by the depletion protocol [46] [1].
- qPCR Measurement: Quantify host and bacterial DNA loads before and after depletion to calculate efficiency and bacterial retention rates objectively [46] [2] [47].
Host Depletion Method Performance Comparison
The performance of host depletion methods varies significantly by sample type and specific metric. The table below summarizes key quantitative data from recent studies.
Table 1: Performance of Host Depletion Methods Across Different Sample Types
| Method | Sample Type | Host DNA Reduction (vs. Raw) | Microbial Read Increase (vs. Raw) | Key Advantages / Disadvantages |
|---|---|---|---|---|
| Saponin + Nuclease (S_ase) [46] | Bronchoalveolar Lavage Fluid (BALF) | To 1.1‱ of original (highly efficient) | 55.8-fold | High host depletion efficiency; may diminish certain pathogens like Mycoplasma pneumoniae |
| HostZERO (K_zym) [46] [2] [47] | BALF / Tissue (DFI) | To 0.9‱ of original / 57-fold reduction in 18S/16S ratio | 100.3-fold (BALF) | Consistently high efficiency and increased bacterial DNA proportion; lower bacterial DNA retention in some BALF samples |
| QIAamp Microbiome (K_qia) [46] [2] [47] | BALF / Sputum / Tissue (DFI) | 32-fold reduction in 18S/16S ratio | 55.3-fold (BALF); 25-fold (Sputum) | Good host depletion and bacterial retention; may alter Gram-negative bacteria proportions in sputum |
| MolYsis [2] | Sputum | 69.6% decrease in host read proportion | 100-fold | Very effective for sputum; may increase final read count significantly |
| Filtration + Nuclease (F_ase) [46] | BALF | - | 65.6-fold | Balanced performance with lower taxonomic bias |
| Novel ZISC Filtration [1] | Whole Blood | >99% WBC removal | >10-fold (vs. unfiltered gDNA) | Excellent for blood; preserves microbial composition; enables gDNA-based mNGS |
Detailed Experimental Protocols
Protocol 1: Evaluating Host Depletion Efficiency in Respiratory Samples
This protocol is adapted from studies benchmarking host depletion methods using bronchoalveolar lavage fluid (BALF) and oropharyngeal swabs [46] [2].
1. Sample Preparation and Pre-Processing:
- Fresh Sample Handling: For BALF, centrifuge at low speed to pellet cells. Resuspend the pellet in an appropriate buffer for downstream processing.
- Frozen Sample Consideration: If samples were frozen without cryoprotectant, note that viability of some bacteria (e.g., Pseudomonas aeruginosa) may be reduced. The addition of 25% glycerol as a cryoprotectant prior to freezing can mitigate this [46] [2].
- Cell-Free DNA Awareness: A significant portion (over 68% in BALF) of microbial DNA may be cell-free. Pre-extraction methods will remove this DNA, affecting overall microbial recovery [46].
2. Host Depletion Treatment (Example Methods):
- Saponin Lysis + Nuclease (S_ase): Treat sample with 0.025% saponin to lyse human cells, followed by nuclease digestion to degrade exposed host DNA. Centrifuge to pellet intact microbial cells [46].
- Commercial Kits (e.g., HostZERO, QIAamp): Follow manufacturer's instructions. These often combine selective lysis of human cells with nuclease treatment or differential binding [46] [2] [47].
3. DNA Extraction and Quality Control:
- Extract DNA from the post-depletion sample using a robust microbial DNA extraction kit.
- Quantitative QC: Use qPCR with primers targeting a single-copy human gene (e.g., RNase P) and a bacterial gene (e.g., 16S rRNA) to calculate:
4. Library Preparation and Sequencing:
- Prepare sequencing libraries from an equal amount of DNA (e.g., 100 ng) from both raw and depleted samples.
- Sequence on an Illumina or MGI platform to a sufficient depth (e.g., 10-20 million reads per sample is a common baseline, but deeper sequencing is beneficial) [46] [1].
5. Bioinformatic Analysis and Metric Calculation:
- Remove adapter sequences and low-quality bases from raw sequencing reads.
- Align reads to the human reference genome (e.g., hg38) using tools like Bowtie2 to identify and remove host-derived reads.
- Align non-host reads to a comprehensive microbial genome database.
- Calculate the final metrics:
Protocol 2: Application for Diagnosing Pulmonary Tuberculosis (HDA-mNGS)
This optimized protocol for detecting Mycobacterium tuberculosis highlights a clinical application [16].
1. Sample Pre-Treatment:
- Add a mucolytic agent like Sputasol to BALF samples. Vortex and incubate at room temperature for 15 minutes to homogenize the sample.
2. Host Depletion and DNA Extraction:
- Centrifuge the treated sample to pellet cells.
- Resuspend the pellet and apply a saponin-based host depletion method to lyse human cells and digest host DNA.
- Extract DNA from the remaining intact microbial cells using a commercial kit (e.g., TIANamp Micro DNA Kit).
3. Downstream Analysis:
- Proceed with library construction and sequencing (e.g., on MGISEQ-2000).
- For a rapid turnaround, host-depleted DNA can also be used with nanopore sequencing (HDA-Nanopore).
- In the bioinformatic analysis, pay particular attention to the increase in MTB genome coverage and the improved depth for detecting antimicrobial resistance (AMR) loci, which are key indicators of success [16].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Host Depletion Experiments
| Reagent / Kit Name | Function / Principle | Sample Type Applicability |
|---|---|---|
| Saponin [46] | Detergent that selectively lyses mammalian cells without disrupting bacterial cell walls. | Respiratory samples (BALF, sputum), other high-host content samples. |
| Benzonase Nuclease [2] | Degrades DNA in the solution after host cell lysis, leaving intracellular microbial DNA protected. | Sputum, skin swabs, saliva. |
| HostZERO Microbial DNA Kit [46] [2] [47] | Commercial kit using selective lysis and nuclease treatment to remove host DNA. | Tissue, BALF, sputum. Consistently shows high efficiency. |
| QIAamp DNA Microbiome Kit [46] [2] [47] | Commercial kit using enzymatic lysis and column-based separation to enrich microbial DNA. | Tissue, BALF, sputum. Good balance of efficiency and DNA retention. |
| Novel ZISC-based Filtration Device [1] | Filter that physically removes host white blood cells while allowing microbes to pass through. | Whole blood. Integrates with gDNA-based mNGS workflows. |
| MolYsis Basic Kit [2] | Commercial kit series using a multi-step enzymatic and binding protocol to remove host DNA. | Sputum, nasal swabs. |
| ZymoBIOMICS Microbial Community Standard [1] | Defined mock community of bacterial and fungal species. Serves as a positive control to assess bias and contamination. | All sample types (spiked into sample or processed separately). |
Troubleshooting Guide: Common Issues in Host Depletion
FAQ: Addressing Microbial Loss During Host DNA Removal
1. Why is my microbial DNA yield low after host depletion, and how can I improve it? Low microbial yield after host depletion is often due to method-induced cell loss or damage. To improve recovery:
- Evaluate Bacterial Retention: Choose a method that minimally impacts microbial integrity. For example, novel filtration techniques like the ZISC-based filter have demonstrated >99% white blood cell removal while allowing unimpeded passage of bacteria and viruses [1].
- Avoid Harsh Lysis Conditions: When using methods that lyse host cells, ensure the concentration of chemical agents like saponin is optimized (e.g., as low as 0.025%) to reduce collateral damage to susceptible microbes [46].
- Assess Microbial Retention Rates: Refer to performance data for different methods. In one study, the nuclease digestion (Rase) method showed the highest bacterial DNA retention rate in BALF samples (median 31%), whereas osmotic lysis followed by PMA treatment (Opma) showed poor microbial recovery [46].
- Verify Input Sample Quality: Ensure that the input sample is fresh and processed correctly. Using old blood samples (e.g., older than one week) or improper storage can lead to inherent DNA degradation and loss of yield [48].
2. How does the host depletion method alter the apparent microbial community composition? Host depletion methods can introduce taxonomic bias by disproportionately affecting certain microorganisms.
- Identify Vulnerable Taxa: Be aware that some commensals and pathogens, including Prevotella spp. and Mycoplasma pneumoniae, can be significantly diminished by certain host depletion procedures [46].
- Select a Balanced Method: Benchmarking studies reveal that methods perform differently. For instance, the filtration with nuclease digestion (F_ase) method demonstrated one of the most balanced performances in preserving microbial composition, while other methods introduced more significant alterations in microbial abundance [46].
- Use Mock Communities: For critical compositional studies, validate your chosen host depletion workflow using a mock microbial community with known abundances to quantify the bias introduced [46].
3. My sequencing results show high host read counts even after depletion. What went wrong? This indicates inefficient host DNA removal. The causes and solutions include:
- Check Depletion Efficiency: Confirm the method's capability for your sample type. For example, methods like saponin lysis with nuclease (Sase) and the HostZERO kit (Kzym) have shown high host DNA removal efficiency, reducing host load by up to four orders of magnitude in respiratory samples [46].
- Optimize Experimental Conditions: For methods involving saponin, the concentration is critical. Test and optimize conditions for your specific samples, as concentrations used in literature can vary from 0.025% to 2.50% [46].
- Consider Sample Type: The effectiveness of some methods depends on the sample matrix. Post-extraction methods that remove methylated host DNA (e.g., with NEBNext Microbiome DNA Enrichment Kit) have shown poor performance in respiratory samples, consistent with findings from other sample types [46].
Performance Comparison of Host Depletion Methods
The following table summarizes the performance of various host depletion methods as benchmarked in respiratory samples, providing a guide for expected outcomes [46].
Table 1: Benchmarking of Host Depletion Methods in Respiratory Samples
| Method (Abbreviation) | Description | Host DNA Removal Efficiency | Bacterial DNA Retention in BALF | Key Considerations |
|---|---|---|---|---|
| Saponin + Nuclease (S_ase) | Lysis of human cells with saponin, followed by digestion of freed DNA. | Very High (to 0.01% of original) | Low | High host removal but can reduce bacterial load. |
| HostZERO Kit (K_zym) | Commercial kit for host cell lysis and DNA degradation. | Very High (to 0.01% of original) | Low | Similar profile to S_ase. |
| Filtration + Nuclease (F_ase) | Physical filtration to remove host cells, followed by nuclease treatment. | High | Moderate | Balanced performance, minimal taxonomic bias. |
| QIAamp Microbiome Kit (K_qia) | Commercial kit using differential lysis. | Moderate | High (in OP samples) | Good bacterial retention but lower host removal. |
| Nuclease Digestion (R_ase) | Digestion of extracellular, cell-free DNA. | Low | High (Median 31%) | Preserves bacteria well but leaves intracellular host DNA. |
| Osmotic Lysis + Nuclease (O_ase) | Hypotonic lysis of human cells followed by nuclease digestion. | Moderate | Moderate | - |
| Osmotic Lysis + PMA (O_pma) | Hypotonic lysis followed by PMA degradation of DNA. | Low | Low | Least effective for increasing microbial reads. |
Experimental Protocols for Key Methodologies
Detailed Protocol: ZISC-Based Filtration for Blood Samples
This protocol is adapted from a study optimizing mNGS for sepsis diagnosis, which achieved >99% white blood cell removal and a tenfold enrichment of microbial reads [1].
1. Sample Preparation:
- Collect whole blood using EDTA as an anticoagulant. Do not use heparin, as it inhibits downstream PCR reactions [49].
- Process samples fresh or store them appropriately at 4°C for short periods to prevent white blood cell degradation. Avoid multiple freeze-thaw cycles.
2. Host Cell Depletion Filtration:
- Transfer a defined volume of whole blood (e.g., 3-13 mL) into a syringe.
- Securely attach the novel ZISC-based fractionation filter (e.g., Devin filter from Micronbrane) to the syringe.
- Gently depress the syringe plunger to pass the blood sample through the filter into a clean 15 mL collection tube.
- The ZISC coating selectively binds and retains host leukocytes while allowing bacteria and viruses to pass through unimpeded [1].
3. Separation of Microbial Pellet:
- Centrifuge the filtered blood at low speed (e.g., 400g for 15 minutes) to isolate plasma.
- Transfer the plasma to a new tube and perform a high-speed centrifugation (e.g., 16,000g) to obtain a microbial cell pellet.
4. DNA Extraction and Library Preparation:
- Extract genomic DNA (gDNA) from the microbial pellet using a standard microbial DNA extraction kit.
- Proceed with library preparation for mNGS. The cited study used the Ultra-Low Library Prep Kit and sequenced on an Illumina NovaSeq 6000, aiming for at least 10 million reads per sample [1].
Detailed Protocol: Filtration with Nuclease (F_ase) for Respiratory Samples
This method was developed and benchmarked against other techniques for BALF and oropharyngeal swab samples, showing a balanced performance with high microbial read enrichment (65.6-fold) [46].
1. Sample Pre-treatment:
- Add a cryoprotectant like 25% glycerol to the respiratory sample (BALF or OP swab in solution) to preserve microbial integrity during processing [46].
2. Host Cell Removal by Filtration:
- Pass the sample through a 10 μm filter. This pore size is designed to retain larger human cells while allowing most bacterial and viral particles to pass through.
3. Digest Residual Host DNA:
- Treat the filtrate with a nuclease enzyme to digest any residual host DNA that may have been released from lysed cells during filtration.
4. Microbial DNA Extraction:
- Concentrate the microorganisms from the nuclease-treated filtrate via centrifugation.
- Extract the total DNA from the resulting pellet for downstream mNGS analysis.
Workflow and Decision Pathways
Host Depletion Method Selection Guide
This diagram illustrates a logical pathway for selecting an appropriate host depletion method based on key experimental goals and sample types.
Experimental Workflow for gDNA-based mNGS with Host Depletion
This diagram outlines the core steps for processing a blood sample using a genomic DNA-based workflow that incorporates a host depletion step, proven to significantly enhance pathogen detection in sepsis [1].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Key Reagents and Kits for Host Depletion and Microbial DNA Recovery
| Reagent/Kit Name | Function | Key Features and Considerations |
|---|---|---|
| ZISC-Based Filtration Device (e.g., Devin filter) | Pre-extraction physical removal of host white blood cells from whole blood. | >99% WBC removal; preserves microbial integrity; suitable for gDNA-based mNGS from blood [1]. |
| QIAamp DNA Microbiome Kit (Qiagen) | Pre-extraction method using differential lysis to remove host cells. | More efficient than no depletion; less labor-intensive than some methods; performance varies by sample type [1] [46]. |
| HostZERO Microbial DNA Kit (Zymo Research) | Pre-extraction chemical lysis of host cells and degradation of host DNA. | Very high host DNA removal efficiency; may reduce bacterial load and alter community composition [46]. |
| NEBNext Microbiome DNA Enrichment Kit (New England Biolabs) | Post-extraction method that removes CpG-methylated host DNA. | Can be inefficient for samples with very high host DNA background, such as respiratory samples [1] [46]. |
| Saponin | Detergent for selectively lysing mammalian cells in pre-extraction methods. | Effectiveness is concentration-dependent; low concentrations (e.g., 0.025%) are recommended to minimize microbial loss [46]. |
| Nuclease Enzymes (e.g., DNase) | Digests DNA in solution, typically used after host cell lysis or filtration to remove free host DNA. | Critical for removing host DNA from lysates or filtrates; does not affect DNA within intact microbial cells [46]. |
In chemogenomic Next-Generation Sequencing (NGS) research, the overwhelming presence of host DNA in samples poses a significant technical challenge. It can consume over 90% of sequencing reads, drastically reducing the depth of microbial data and compromising the sensitivity of your experiments [4]. This technical support center provides targeted troubleshooting guides and FAQs to help you navigate the critical variables of sample processing—volume, storage conditions, and matrix effects—to effectively reduce host DNA background and ensure the success of your NGS workflows.
Frequently Asked Questions (FAQs) and Troubleshooting Guides
FAQ 1: How does sample volume impact host DNA depletion efficiency?
- Question: "I process different sample volumes in my lab. Will this affect how well I can remove host DNA?"
- Answer: Yes, sample volume is a critical variable. The efficiency of physical depletion methods, like filtration, must be validated across the volume range you use. One study demonstrated that a novel filtration device efficiently removed host white blood cells (>99%) across a range of blood volumes (3mL to 13mL) while allowing microbes to pass through [1]. Always ensure your chosen method is optimized for your standard operating volumes.
FAQ 2: What is the quantitative impact of host DNA on my sequencing results?
- Question: "My sequencing run had a high percentage of human reads. How much has this affected my data?"
- Answer: The impact is severe and quantifiable. Research shows that as the proportion of host DNA increases, the sensitivity for detecting microbial species drops significantly. The table below summarizes the correlation between host DNA levels and sequencing efficacy.
| Host DNA in Sample | Impact on Microbial Detection |
|---|---|
| 90% Host DNA | Major impact on sensitivity; leads to an increased number of undetected species, especially with reduced sequencing depth [4]. |
| 99% Host DNA | Microbiome profiling becomes highly inaccurate and inconsistent, making it difficult to obtain meaningful microbial data [4]. |
FAQ 3: My NGS library yield is low after host depletion. What went wrong?
- Question: "I used a host DNA depletion method, but my final library concentration is much lower than expected. What are the main causes?"
- Answer: Low library yield after depletion can stem from several issues in the preparation workflow. The following table outlines common culprits and their solutions.
| Root Cause | Mechanism of Yield Loss | Corrective Action |
|---|---|---|
| Overly aggressive purification/size selection | Desired microbial DNA fragments are accidentally discarded during clean-up steps [21]. | Optimize bead-to-sample ratios and size selection parameters to maximize recovery of target fragments. |
| Poor input DNA quality / contaminants | Residual salts or organics inhibit enzymes in downstream ligation or amplification steps [21]. | Re-purify the input sample, ensure high purity (260/280 ~1.8), and use fresh wash buffers. |
| Inaccurate quantification / pipetting error | Suboptimal enzyme stoichiometry due to inaccurate DNA concentration measurements [21]. | Use fluorometric quantification (e.g., Qubit) instead of UV absorbance; calibrate pipettes; use master mixes to reduce volumetric errors. |
FAQ 4: Are there specific storage conditions to preserve sample integrity for host DNA depletion?
- Question: "How should I store my samples before processing to ensure successful host DNA depletion later?"
- Answer: While the search results do not specify exact storage conditions for host DNA depletion, best practices for preserving nucleic acid integrity are paramount.
- Fresh is best: Fresh starting material is always recommended for optimal results [50].
- Proper preservation: If immediate processing is not possible, samples should be stored appropriately, typically by freezing at specific temperatures to prevent DNA degradation [50].
- Avoid degradation: Using degraded DNA can lead to low-quality sequencing data and complicate depletion efficiency [51].
Experimental Protocols: Key Methodologies
Protocol 1: ZISC-Based Filtration for Host Cell Depletion
This protocol details a method for pre-extraction host cell depletion from whole blood, leveraging a novel zwitterionic interface coating [1].
- Sample Preparation: Collect whole blood using standard phlebotomy techniques. For validation, samples can be spiked with known microbial communities.
- Filtration Setup: Securely connect a novel ZISC-based fractionation filter (e.g., the "Devin" filter from Micronbrane) to a syringe.
- Host Cell Depletion: Transfer a defined volume of whole blood (e.g., 4 mL) into the syringe. Gently depress the plunger to pass the blood through the filter into a clean collection tube. The filter selectively binds and retains host leukocytes.
- Microbial Pellet Isolation: Centrifuge the filtered blood at low speed (e.g., 400g for 15 min) to isolate plasma. Then, perform a high-speed centrifugation (e.g., 16,000g) of the plasma to obtain a pellet containing microbial cells.
- DNA Extraction: Proceed with genomic DNA (gDNA) extraction from the microbial pellet using a standard kit.
Protocol 2: Differential Lysis for Host DNA Depletion in Tissue Biopsies
This protocol describes a method to remove host DNA from tissue biopsies, such as colon tissue, by exploiting the differential fragility of mammalian and bacterial cells [52].
- Tissue Processing: Human or mouse colon biopsies are collected and divided into groups for treatment and control.
- Differential Lysis: Subject the biopsy samples to a lysis buffer and conditions designed to lyse mammalian cells while leaving bacterial cells intact.
- DNase Treatment (Optional): Add a DNase enzyme to degrade the released host DNA. The DNase must subsequently be inactivated before bacterial lysis.
- Bacterial Lysis: Apply a stronger lysis buffer (e.g., involving bead-beating or enzymatic digestion) to break open the bacterial cells and release microbial DNA.
- DNA Purification: Purify the total DNA, which is now enriched for microbial content, using a commercial kit.
- Validation: The effectiveness of depletion is measured by comparing the percentage of host and bacterial reads in sequenced treated samples versus non-depleted controls.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Kit | Function in Host DNA Depletion |
|---|---|
| ZISC-Based Filtration Device | Physically removes host white blood cells from liquid samples like blood, preserving microbes for downstream gDNA extraction [1]. |
| QIAamp DNA Microbiome Kit | Uses differential lysis to selectively remove human cells, enriching for microbial DNA [1]. |
| NEBNext Microbiome DNA Enrichment Kit | Employes post-extraction enzymatic removal of CpG-methylated host DNA [1]. |
| Nextera XT DNA Library Prep Kit | Used for preparing sequencing libraries from metagenomic DNA after host depletion [4]. |
| Agencourt AMPure XP Beads | Magnetic beads used for post-library preparation clean-up to remove short fragments and purify the final NGS library [4]. |
Workflow Visualization: Standard vs. Host-Depleted NGS
The following diagram illustrates the key differences between a standard NGS workflow and one that incorporates a host DNA depletion step, highlighting the points where sample processing variables are most critical.
Key Takeaways for Your Research
Successful reduction of host DNA background hinges on a holistic approach to sample processing. Key considerations include:
- Validate by Volume: Ensure your chosen host depletion method is effective across the entire range of sample volumes used in your lab [1].
- Prioritize Sample Integrity: Proper handling and storage of samples from collection to processing is fundamental to preserving the true microbial profile and ensuring depletion efficiency [50].
- Understand Matrix Effects: Be aware that the sample matrix itself can introduce biases and interferences at multiple stages. A thorough understanding of your sample type is crucial for troubleshooting [53].
- Choose the Right Tool: Select a depletion strategy (physical, enzymatic, or chemical) that is compatible with your sample type and research objectives [1] [52].
Frequently Asked Questions (FAQs)
FAQ 1: My host depletion method is not effectively reducing human DNA background, leading to poor microbial read counts. What could be wrong and how can I fix it?
Ineffective host DNA depletion is a common bottleneck that consumes sequencing resources and obscures pathogenic signals. The table below summarizes the core problems and validated solutions.
| Problem | Failure Signs | Root Causes | Corrective & Validation Strategies |
|---|---|---|---|
| Inefficient Experimental Depletion | Host DNA still >95% post-depletion; Low microbial RPM (e.g., <1000 RPM) [5] [16]. | Non-optimized filtration; Inefficient lysis of host cells; Method unsuitable for sample type (e.g., using cfDNA for intracellular pathogens) [5] [19]. | - Switch to advanced filters: Use a ZISC-based filtration device, shown to achieve >99% white blood cell removal and a >10x increase in microbial reads [5] [19].- Use gDNA, not cfDNA: For intracellular pathogens like Mycobacterium tuberculosis, genomic DNA (gDNA) from cell pellets is superior to cell-free DNA (cfDNA) for pre-extraction host depletion [19] [16]. |
| Inadequate Computational Filtration | False positive microbial calls; Apparent sex-based biases in microbial profiles [54]. | Using an incomplete human reference genome (e.g., GRCh38) that misses regions like the complete Y chromosome, causing human reads to be misclassified as microbial [54]. | - Upgrade reference genome: Implement a comprehensive human reference like T2T-CHM13v2.0, which includes a complete Y chromosome and abolishes artifactual sex biases [54].- Use "cleaned" databases: Employ microbial databases where regions with human sequence similarity have been masked (e.g., RS210-clean) [54]. |
Experimental Protocol: ZISC-Based Filtration for Blood Samples
This protocol, adapted from Chen et al. (2025), details a robust method for enriching microbial cells from whole blood [5] [19].
- Sample Preparation: Draw 3-13 mL of whole blood into an anti-coagulant tube. For validation, spike in a known quantity of a control organism (e.g., E. coli at 10⁴ CFU/mL).
- Filtration Setup: Securely connect a sterile, novel ZISC-based fractionation filter (e.g., Devin filter) to a syringe.
- Host Cell Depletion: Transfer the blood sample into the syringe. Gently depress the plunger to push the blood through the filter into a clean 15 mL collection tube. The ZISC coating retains host leukocytes while allowing bacteria and viruses to pass unimpeded.
- Microbial Pellet Collection: Centrifuge the filtrate at low speed (400g for 15 min) to isolate plasma. Then, perform a high-speed centrifugation (16,000g) of the plasma to obtain a microbial cell pellet.
- DNA Extraction and Sequencing: Proceed with DNA extraction from the pellet using a standard microbial DNA kit. Prepare libraries and sequence on your preferred NGS platform.
This workflow has been clinically validated to detect all expected pathogens in sepsis samples, boosting microbial read counts from an average of 925 RPM (unfiltered) to 9,351 RPM (filtered) [5].
FAQ 2: My mNGS results show a distorted microbial community profile that doesn't match expected abundances. How do I correct for this bias?
Distorted microbial profiles can arise from both wet-lab and computational biases, misleading biological interpretations. The following table outlines key sources and correction methods.
| Problem | Failure Signs | Root Causes | Corrective & Validation Strategies |
|---|---|---|---|
| Wet-lab Extraction Bias | Inconsistent recovery of taxa across different extraction kits or lysis conditions; Does not correlate with true abundance [55]. | Differential lysis efficiency of bacterial cells due to variations in cell wall structure (e.g., Gram-positive vs. Gram-negative) and morphology [55]. | - Use mock communities: Include a standardized mock community with known abundances in your extraction batch to quantify bias per protocol [55].- Morphology-based correction: Computational correction of extraction bias based on bacterial cell morphology (e.g., size, shape) can significantly improve accuracy, even for non-mock taxa [55]. |
| Bioinformatic Database Bias | Over- or under-representation of specific species; Inflated diversity metrics [54] [56]. | PCR amplification biases from different 16S rRNA regions, polymerases, or sequencing platforms; Mismapping of reads due to sequence homology [56]. | - Apply a reference-based bias correction model: Use a model calibrated with droplet digital PCR (ddPCR) data from mock communities to correct biased sequencing ratios. This works across platforms and 16S regions [56].- Validate with quantitative metrics: Use quantitative diversity metrics (e.g., Weighted UniFrac, Bray-Curtis) which are more sensitive to falsely inflated abundances, and compare results after applying bias correction [54]. |
Experimental Protocol: Saponin-Based Host DNA Depletion for BALF Samples
This protocol is optimized for low-biomass samples like Bronchoalveolar Lavage Fluid (BALF), where intracellular pathogens reside within host cells [16].
- Sample Pre-treatment: Add Sputasol (Oxoid) or a saponin-based solution to the BALF sample. Vortex and incubate at 42°C for 10 minutes. Saponin selectively permeabilizes mammalian (host) cell membranes without lysing robust bacterial cells.
- Differential Centrifugation: Centrifuge the treated sample. The released host DNA remains in the supernatant, while intact microbial cells form a pellet.
- Supernatant Removal: Carefully remove and discard the supernatant containing the bulk of solubilized host DNA.
- Microbial Lysis and DNA Extraction: Resuspend the microbial pellet and proceed with rigorous mechanical and enzymatic lysis to break open the microbial cells. Extract DNA using a microbial DNA kit.
- Sequencing and Analysis: Construct libraries and sequence. This HDA-mNGS method has been shown to increase MTB genome coverage by up to 16-fold and significantly improve diagnostic sensitivity [16].
The Scientist's Toolkit: Essential Research Reagents
| Item | Function | Application Context |
|---|---|---|
| ZISC-based Filtration Device | Physically removes >99% of host white blood cells from whole blood by selective binding, enriching microbial passage [5] [19]. | Host depletion from blood samples for gDNA-based mNGS in sepsis and bloodstream infection research. |
| Saponin Reagent | Selective chemical depletion agent that permeabilizes mammalian host cells without lysing bacterial cells, allowing host DNA washaway [16]. | Host depletion from samples with intracellular pathogens (e.g., BALF for tuberculosis) or low microbial biomass. |
| Mock Microbial Communities | Defined controls with known microbial composition and abundance used to quantify and correct for technical biases across the entire workflow [55] [56]. | Essential for validating extraction protocols, quantifying bias, and calibrating bioinformatic correction models. |
| T2T-CHM13v2.0 Genome | A complete human reference genome that includes previously missing regions (e.g., Y chromosome), preventing human read misclassification [54]. | Critical for comprehensive computational host read filtration to avoid false positives and artifactual biases. |
| rpoB Gene ddPCR Assays | Highly specific, quantitative assays targeting the single-copy rpoB gene for absolute bacterial quantification, independent of 16S copy number variation [56]. | Used to establish ground-truth ratios in mock or complex communities for calibrating reference-based bias correction models. |
FAQs: Addressing Core Challenges in Host DNA Depletion
Q1: What is "host contamination" and why is it a problem in chemogenomic NGS? Host contamination occurs when DNA from the host organism (e.g., human DNA in a blood sample) dominates the sequencing library. This excessive host DNA background consumes sequencing capacity, reduces microbial read depth, and severely compromises the sensitivity for detecting pathogen signals [57] [19].
Q2: My NGS library yield is unexpectedly low after host depletion. What are the primary causes? Low library yield can stem from several issues in the preparation workflow. Common causes include poor input DNA quality, contaminants inhibiting enzymes, inaccurate DNA quantification, suboptimal adapter ligation, or overly aggressive purification and size selection that leads to sample loss [21].
Q3: How can I verify that my host depletion method is working effectively? Effective host depletion is confirmed by both pre- and post-sequencing metrics. Pre-sequencing, use a cell counter to measure white blood cell (WBC) removal; efficient methods should achieve >99% WBC depletion [5] [19]. Post-sequencing, calculate the percentage of sequencing reads that align to the host genome; a significant reduction indicates successful depletion.
Q4: Are computational methods sufficient to correct for high host DNA background in sequencing data? While computational subtraction of host reads can help, it is not a complete solution. It recovers sequencing capacity but cannot rescue microbial reads lost during wet-lab preparation due to low initial abundance. A combined approach of wet-lab depletion to physically remove host DNA, followed by computational cleaning, is most effective [5] [19].
Q5: What are the advantages of genomic DNA (gDNA)-based mNGS with host depletion over cell-free DNA (cfDNA)-based mNGS? gDNA-based mNGS coupled with wet-lab host depletion enables physical enrichment of intact microbial cells before DNA extraction. This approach has demonstrated over a tenfold increase in microbial reads and 100% detection of expected pathogens in culture-positive sepsis samples, outperforming cfDNA-based methods which show inconsistent sensitivity [19].
Troubleshooting Guide: Host DNA Background and Sequencing Failures
Table: Common NGS Preparation Problems in Host DNA Depletion Studies
| Problem Category | Typical Failure Signals | Common Root Causes | Corrective Actions |
|---|---|---|---|
| Sample Input / Quality | Low yield; low library complexity; smear in electropherogram | Degraded DNA; sample contaminants (salts, phenol); inaccurate quantification | Re-purify input; use fluorometric quantification (Qubit); check 260/230 and 260/280 ratios [21] |
| Fragmentation & Ligation | Unexpected fragment size; sharp ~70-90 bp peak (adapter dimers) | Over/under-shearing; improper adapter-to-insert molar ratio; poor ligase performance | Optimize fragmentation parameters; titrate adapter concentration; ensure fresh enzymes and buffers [21] |
| Amplification & PCR | High duplicate rate; over-amplification artifacts; bias | Too many PCR cycles; carryover enzyme inhibitors; mispriming | Reduce cycle number; use hot-start polymerase; optimize annealing temperature; add GC enhancer for difficult templates [21] [58] |
| Purification & Cleanup | Adapter dimer carryover; high background; sample loss | Wrong bead-to-sample ratio; over-drying beads; inefficient washing | Precisely follow cleanup protocols; avoid bead over-drying; use master mixes to reduce pipetting errors [21] |
Table: Troubleshooting Low NGS Library Yield
| Cause of Low Yield | Mechanism of Yield Loss | Corrective Action |
|---|---|---|
| Poor Input Quality / Contaminants | Enzyme inhibition during fragmentation or ligation. | Re-purify sample; ensure wash buffers are fresh; target 260/230 > 1.8 [21] |
| Inaccurate Quantification | Suboptimal enzyme stoichiometry due to over/under-estimated DNA. | Use fluorometric methods (Qubit) over UV absorbance; calibrate pipettes [21] |
| Suboptimal Adapter Ligation | Reduced adapter incorporation into library fragments. | Titrate adapter:insert ratio; use fresh ligase/buffer; optimize incubation [21] |
| Overly Aggressive Size Selection | Desired library fragments are accidentally discarded. | Optimize bead-to-sample ratio; avoid over-drying beads during cleanup [21] |
Experimental Protocols: Key Methodologies
Protocol: ZISC-Based Filtration for Host Cell Depletion
This protocol details a novel wet-lab method for depleting host white blood cells to enhance pathogen detection in blood samples [5] [19].
- Principle: A Zwitterionic Interface Ultra-Self-assemble Coating (ZISC)-based filter selectively binds and retains host leukocytes while allowing bacteria and viruses to pass through unimpeded.
- Materials:
- Devin filter (Micronbrane, Taiwan)
- Whole blood sample (3-13 mL volumes tested)
- Syringe
- 15 mL Falcon tube
- Low-speed and high-speed centrifuges
- Procedure:
- Transfer approximately 4 mL of whole blood into a syringe.
- Securely connect the ZISC-based filter to the syringe.
- Gently depress the syringe plunger to push the blood sample through the filter into a 15 mL Falcon tube.
- Centrifuge the filtered blood at 400g for 15 minutes at room temperature to isolate plasma.
- Subject the plasma to high-speed centrifugation at 16,000g to obtain a microbial pellet.
- Proceed with DNA extraction from the pellet using a standard microbial DNA enrichment kit.
- Performance Validation: This method achieves >99% white blood cell removal and results in a tenfold increase in microbial reads per million (RPM) in downstream mNGS compared to unfiltered samples [19].
Protocol: Computational Detection of Sequence Contamination
This in-silico protocol identifies common contaminants (e.g., vector, adapter sequences) in sequencing data before analysis [59].
- Principle: Using sequence similarity search tools like BLAST against specialized databases of common contaminants.
- Materials:
- Raw sequencing reads (FASTA/FASTQ format)
- NCBI's VecScreen tool and UniVec database
- BLAST suite
- Optional: Webcutter for restriction site analysis
- Procedure for Vector Contamination:
- Access the NCBI VecScreen tool.
- Input your nucleotide sequence in FASTA format.
- VecScreen runs a BLAST search against the UniVec database and categorizes matches.
- Review the graphical output to identify the location and strength of contaminating segments.
- Procedure for Adapter/Linker Contamination:
- Perform a nucleotide BLAST (blastn) search.
- Use the specific oligonucleotide sequences of the adapters or primers used in your library prep as the query sequence.
- Set your sequencing reads as the database to search against.
- Significant hits indicate adapter contamination, necessitating trimming before analysis.
- Note: Always screen sequences before submission to public databases or downstream analysis to avoid erroneous conclusions and wasted resources [59].
Diagram Title: Integrated Wet-lab and Computational Workflow for NGS
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Reagents for Host DNA Depletion and NGS
| Item | Function | Example Use-Case |
|---|---|---|
| Devin Filter (ZISC-based) | Physically depletes host white blood cells from whole blood via surface coating that binds leukocytes. | Pre-extraction host depletion in gDNA-based mNGS workflows for sepsis [5] [19]. |
| QIAamp DNA Microbiome Kit | Uses differential lysis to selectively remove human host DNA from samples. | An alternative method for host DNA depletion [19]. |
| NEBNext Microbiome DNA Enrichment Kit | Enriches microbial DNA by selectively binding and removing methylated host (human) DNA. | Post-extraction host DNA depletion [19]. |
| High-Fidelity DNA Polymerase | Reduces PCR errors during library amplification; essential for complex or GC-rich templates. | Library amplification in NGS; e.g., Q5 High-Fidelity Polymerase [58]. |
| PCR Cleanup Kits | Remove excess salts, primers, and adapter dimers post-amplification to reduce background. | Purification after library amplification and size selection [21]. |
| UniVec Database | A curated database of vector and adapter sequences used for in-silico contamination screening. | Identifying and removing contaminating sequences from NGS data using VecScreen [59]. |
Diagram Title: Systematic Troubleshooting for Low NGS Yield
Frequently Asked Questions (FAQs)
FAQ 1: What are the primary consequences of high host DNA background in my NGS data? High host DNA background consumes a large portion of your sequencing reads, severely reducing the sensitivity for detecting pathogen or microbial signals. This leads to lower coverage of the target microbiomes, potentially missing low-abundance pathogens, and increases sequencing costs as more depth is required to achieve meaningful results [60].
FAQ 2: Beyond commercial kits, what are some fundamental sample preparation errors that increase host background? Common errors include inaccurate quantification of input DNA, using degraded nucleic acid templates, and the presence of contaminants like phenol, salts, or ethanol that inhibit enzymatic reactions during library preparation [21] [61]. Proper purification and using fluorometric quantification (e.g., Qubit) over absorbance methods are critical [21] [62].
FAQ 3: My library yield is low after host DNA depletion. What should I investigate? Low yield can result from overly aggressive purification, sample loss during manual handling steps, or suboptimal adapter ligation due to improper molar ratios [21]. Ensure you are using the correct bead-based cleanup ratios and verify the quality and concentration of your input material immediately before library prep [21] [61].
Troubleshooting Guide: Common Issues and Solutions
The following table summarizes common problems, their root causes, and corrective actions for managing host DNA background and ensuring library quality [21].
| Problem & Symptoms | Root Cause | Corrective Action |
|---|---|---|
| Low Library Yield• Low final concentration• Broad/faint electropherogram peaks | • Sample loss during manual cleanup steps [21]• Overly aggressive size selection [21]• Enzyme inhibition from contaminants [21] [62] | • Re-purify input DNA to remove inhibitors [62]• Titrate and optimize bead-to-sample ratios during cleanup [21]• Use master mixes to reduce pipetting errors [21] |
| High Host DNA Background• Low on-target rate• Poor pathogen coverage | • Inefficient host DNA depletion method [60]• High abundance of host nucleic acids in low-biomass samples [60] | • Optimize host depletion protocols (e.g., probe-based) [60]• Incorporate robotic liquid handling for consistency [61] |
| Adapter Dimer Contamination• Sharp ~70-90 bp peak on Bioanalyzer | • Inefficient ligation [21]• Suboptimal adapter-to-insert molar ratio [21]• Incomplete cleanup post-ligation [21] | • Titrate adapter concentrations [21]• Optimize bead cleanup parameters to remove short fragments [21] |
| Uneven Coverage / Batch Effects• Inconsistencies across sample batches | • Primer mispriming or bias [61]• Variations in reagents or operators [21] | • Randomize sample processing across batches [61]• Use high-quality, specific primers and automate workflows [61] |
Experimental Protocols for Host DNA Reduction
Protocol 1: Optimizing Bead-Based Cleanup for Size Selection
Bead-based cleanup is critical for removing adapter dimers and selecting the desired insert size, but it is a common point of sample loss.
Detailed Methodology:
- Determine Ratio: Confirm the optimal sample-to-bead ratio (e.g., 0.6X to 0.8X) for your target size selection. Using the wrong ratio can exclude desired fragments or fail to remove small ones [21].
- Mixing and Incubation: Combine beads and sample thoroughly by pipetting. Incubate at room temperature for the recommended time, typically 5-15 minutes [21].
- Pellet on Magnet: Place the tube on a magnetic stand until the solution clears. Carefully remove and discard the supernatant without disturbing the bead pellet [21].
- Washing: While the tube is on the magnet, add fresh 80% ethanol to wash the pellet. Incubate for 30 seconds, then remove the ethanol completely. Avoid over-drying the beads, as a cracked, matte pellet is difficult to resuspend and leads to inefficient elution [21].
- Elution: Remove the tube from the magnet and elute the DNA in a low-EDTA TE buffer or nuclease-free water. Resuspend the beads thoroughly and incubate at room temperature for 2-5 minutes [21].
- Final Separation: Return the tube to the magnet. Once clear, transfer the supernatant containing the purified library to a new tube.
Protocol 2: Automated Library Preparation to Minimize Human Error
Automating the library prep process significantly improves reproducibility and reduces errors related to manual pipetting [61].
Detailed Methodology:
- Platform Selection: Use a liquid handling workstation (e.g., Tecan Fluent, Opentrons OT-2) that is compatible with your chosen library prep kit [63] [61].
- Liquid Class Calibration: Ensure the liquid classes for enzymes, beads, and buffers are correctly calibrated on the robot to ensure accurate and precise liquid transfer [63].
- Utilize Master Mixes: Prepare master mixes of common reagents off-deck to minimize the number of individual pipetting operations the robot must perform [21].
- Include QC Checkpoints: Integrate real-time quality control checks. For instance, AI-powered vision systems can be used with the robot to confirm the presence of pipette tips and correct liquid volumes in wells before proceeding [63].
- Post-Run Recovery: Use "waste plates" to temporarily catch discarded supernatant. This allows for manual retrieval in case of a confirmed error during the automated run [21].
Workflow Visualization for Host DNA Management
The diagram below illustrates the key decision points and strategies for reducing host DNA background in a typical NGS workflow.
Research Reagent Solutions
The table below lists key reagents and materials essential for successful NGS library preparation with minimal host background.
| Item | Function & Rationale |
|---|---|
| Fluorometric Quantification Kits (Qubit) | Accurately measures double-stranded DNA concentration without interference from common contaminants like salts or RNA, preventing inaccurate input material dosing [21] [62]. |
| DNA Depletion Kits (Probe-based) | Selectively removes abundant host (e.g., human) DNA through hybridization capture, dramatically increasing the relative proportion of microbial reads for sequencing [60]. |
| Bead-Based Cleanup Kits (e.g., AMPure XP) | Purifies and size-selects nucleic acid fragments after enzymatic reactions; critical for removing primer dimers and short artifacts [21]. |
| Automated Liquid Handling Platforms | Robotic systems (e.g., Tecan Fluent) perform highly reproducible pipetting, drastically reducing human error and batch effects in high-throughput workflows [63] [61]. |
| Normalized Library Prep Kits | Kits with built-in normalization properties help achieve consistent read depths across different samples, simplifying the workflow and improving data uniformity [61]. |
Performance Validation: Comparative Analysis of Host Depletion Methods
In chemogenomic Next-Generation Sequencing (NGS) research, the overwhelming presence of host DNA presents a fundamental barrier to analytical sensitivity. High levels of host nucleic acids in samples like blood or bronchoalveolar lavage fluid (BALF) consume sequencing resources, effectively masking microbial signals and reducing the detection of pathogenic organisms [1] [16]. This technical challenge directly impacts the Limit of Detection (LOD) and microbial recovery rates, potentially leading to false negatives in pathogen identification. This guide provides troubleshooting and methodological frameworks for researchers seeking to overcome these limitations through host DNA depletion techniques and optimized workflows.
FAQs: Host DNA Depletion and Sensitivity Enhancement
What is the fundamental impact of host DNA on NGS sensitivity?
Host DNA dramatically reduces NGS sensitivity by consuming sequencing capacity. In standard metagenomic NGS (mNGS) of blood samples, human DNA can constitute over 95% of sequenced material, leaving minimal reads for pathogen detection [1] [16]. This background noise elevates the effective Limit of Detection, requiring higher pathogen concentrations for reliable identification. Host DNA depletion methods address this by selectively removing human nucleic acids before sequencing, thereby enriching microbial content and improving detection sensitivity for rare pathogens.
How much sensitivity improvement can host depletion methods provide?
Studies demonstrate significant improvements with optimized host depletion. In sepsis diagnostics, a novel Zwitterionic Interface Ultra-Self-assemble Coating (ZISC)-based filtration device achieved >99% white blood cell removal, resulting in over tenfold enrichment of microbial reads (increasing from 925 to 9,351 reads per million) in clinical samples [1]. Similarly, for pulmonary tuberculosis diagnosis, host DNA depletion-assisted mNGS (HDA-mNGS) improved sensitivity from 51.2% to 72.0% compared to conventional mNGS in BALF samples [16].
Does increasing sequencing depth compensate for high host DNA background?
Evidence suggests that simply increasing sequencing depth is an inefficient strategy for overcoming high host DNA background. One study found that improving sequencing depth did not show a positive effect on improving the detection sensitivity of SARS-CoV-2 in swab samples [7]. Instead, pre-sequencing host DNA removal more effectively enhances sensitivity without the substantial cost increases associated with deeper sequencing.
What are the primary methodological approaches to host DNA depletion?
The two main strategic approaches are:
- Pre-extraction methods: Physically separate host cells from microorganisms before DNA extraction using techniques like filtration [1] or saponin-based chemical lysis [16].
- Post-extraction methods: Remove host DNA after nucleic acid extraction using techniques like CpG-methylated DNA enrichment [1] or DNAse treatment [7].
Table: Comparison of Host DNA Depletion Techniques
| Method | Mechanism | Efficiency | Advantages | Limitations |
|---|---|---|---|---|
| ZISC-based Filtration [1] | Pre-extraction; physical separation | >99% WBC removal | Preserves microbial integrity; high efficiency | Specialized equipment required |
| Saponin-based Depletion [16] | Pre-extraction; chemical lysis | Significant host DNA reduction | Cost-effective; compatible with various samples | Potential impact on some microbial cells |
| Differential Lysis [1] | Pre-extraction; selective lysis | Variable | Commercially available kits | Lower efficiency compared to novel methods |
| CpG-methylated DNA Removal [1] | Post-extraction; enzymatic | Moderate | Works with extracted DNA | May affect microbes with methylated genomes |
| DNAse Treatment [7] | Post-extraction; enzymatic | High for DNA targets | Specific to DNA; preserves RNA | Not suitable for DNA pathogen detection |
Troubleshooting Guides: Solving Common Sensitivity Issues
Problem: Consistently Low Microbial Recovery Despite Host Depletion
Symptoms:
- Lower-than-expected microbial read counts after host depletion
- Inconsistent detection of known spiked controls
- Failure to detect expected pathogens in positive controls
Potential Causes and Solutions:
- Overly aggressive depletion methods damaging microbial cells
Inadequate DNA extraction efficiency for target microbes
- Solution: Incorporate mechanical disruption (bead beating) for tough microbial cell walls; validate extraction efficiency across diverse microbes [64]
Inhibition of downstream enzymatic steps
- Solution: Additional purification steps after host depletion; include inhibition controls in workflow validation [21]
Problem: High Variability in Detection Sensitivity Between Samples
Symptoms:
- Inconsistent LOD between sample batches
- Variable host DNA removal efficiency
- Fluctuating microbial recovery rates
Potential Causes and Solutions:
- Inconsistent sample processing
Variable host cellularity in starting material
- Solution: Normalize input material by cell count or volume; implement pre-analytical sample assessment [1]
Reagent degradation or lot variability
- Solution: Implement strict reagent quality control; test new lots with standardized controls; use master mixes where possible [21]
Symptoms:
- Skewed relative abundances between different microbes
- Consistent under-representation of specific microbial types
- Discrepancy between culture results and sequencing data
Potential Causes and Solutions:
- Differential recovery during host depletion
- Solution: Validate method with diverse microbial spikes; use multiple depletion approaches for comprehensive profiling [1]
Amplification bias in low-input samples
Size-based selection artifacts
- Solution: Minimize fragmentation variations; use size selection methods with high recovery; validate with size standards [21]
Experimental Protocols for Sensitivity Assessment
Protocol 1: Evaluating Host Depletion Efficiency Using Spiked Samples
Purpose: Quantify host DNA removal and microbial recovery rates for method validation [1].
Materials:
- Fresh human whole blood or appropriate biological matrix
- Microbial reference standards (e.g., ZymoBIOMICS D6320 or D6331)
- Host depletion method (filtration, chemical, or enzymatic)
- DNA extraction kit (e.g., TIANamp Micro DNA Kit)
- Fluorometric quantification system (e.g., Qubit fluorometer)
- NGS library preparation reagents
- Bioinformatics pipeline for read classification
Procedure:
- Sample Preparation:
- Aliquot 1-10mL of human whole blood into sterile tubes
- Spike with microbial reference community at known concentrations (e.g., 10²-10⁴ genome equivalents)
- Include unspiked controls for background assessment
Host Depletion:
- Process samples through host depletion method (e.g., ZISC filtration, saponin treatment)
- Retain paired untreated controls for comparison
- Process all samples in replicate (n≥3)
Nucleic Acid Extraction:
- Extract DNA/RNA from both depleted and untreated samples
- Quantify total nucleic acid yield using fluorometric methods
- Assess quality (e.g., fragment analyzer, absorbance ratios)
Library Preparation and Sequencing:
- Prepare sequencing libraries with unique barcodes
- Use consistent input masses across samples
- Sequence on appropriate platform (e.g., Illumina, MGI, Nanopore)
- Generate sufficient coverage (≥10 million reads per sample)
Bioinformatic Analysis:
- Quality filter raw reads (remove adapters, low-quality bases)
- Classify reads as host versus microbial using reference databases
- Calculate: Host DNA removal efficiency, Microbial read recovery, Enrichment factors
Calculation of Key Metrics:
- Host Depletion Efficiency = [1 - (Host readsdepleted / Host readsuntreated)] × 100
- Microbial Recovery = (Microbial readsdepleted / Microbial readsuntreated) × 100
- Enrichment Factor = (Microbial fractiondepleted / Microbial fractionuntreated)
Protocol 2: Limit of Detection (LOD) Determination for NGS Assays
Purpose: Establish the minimum microbial concentration detectable with 99% confidence using the MDL framework [65].
Materials:
- Clean reference matrix (e.g., sterile saline, TE buffer)
- Target microorganism culture or DNA standard
- DNA quantification standard (e.g., synthetic oligo, quantified genomic DNA)
- Full NGS workflow reagents
- Statistical analysis software
Procedure:
- Spike Preparation:
- Prepare dilution series of target microbe in reference matrix
- Span expected detection range (e.g., 10⁰-10⁴ copies/μL)
- Include true negative (unspiked) controls
Sample Processing:
- Process 7-8 replicates per concentration level
- Distribute analysis across multiple batches/days
- Include method blanks with each batch
Data Analysis:
- Calculate mean and standard deviation of microbial reads for each level
- Perform regression of read counts versus input concentration
- Identify the lowest concentration with consistent detection (≥95% detection rate)
Statistical LOD Determination:
- Follow MDL procedure: MDL = t × S, where t is Student's t-value and S is standard deviation [65]
- Use appropriate confidence level (typically 99% for critical detection)
- Verify with independent samples near the calculated LOD
Table: Example LOD Determination for Bacterial Pathogens Using NGS
| Pathogen | Sample Matrix | Host Depletion Method | LOD (Genome Copies) | Sequencing Reads Required |
|---|---|---|---|---|
| Mycobacterium tuberculosis [16] | BALF | Saponin-based | ~10² | ~10 million |
| SARS-CoV-2 [7] | Swab | DNAse treatment | Ct ~35 | 10-20 million |
| Bacterial community [1] | Blood | ZISC filtration | 10² GE | 10 million |
| E. coli/S. aureus [1] | Blood | ZISC filtration | 10⁴ CFU/mL | 5-10 million |
Workflow Visualization: Host DNA Depletion for Enhanced Sensitivity
Host Depletion Enhanced mNGS Workflow: This diagram compares standard and host-depleted metagenomic NGS workflows, highlighting two strategic approaches for host DNA removal that significantly improve detection sensitivity for microbial pathogens.
The Scientist's Toolkit: Essential Reagents and Technologies
Table: Key Research Reagents and Technologies for Sensitivity Optimization
| Category | Specific Product/Technology | Primary Function | Application Notes |
|---|---|---|---|
| Host Depletion Technologies | ZISC-based Filtration Device [1] | >99% WBC removal while preserving microbes | Optimal for blood samples; maintains microbial viability |
| Saponin-based Host Depletion [16] | Selective lysis of human cells | Cost-effective for BALF and sputum samples | |
| QIAamp DNA Microbiome Kit [1] | Differential lysis-based depletion | Commercial solution for various sample types | |
| NEBNext Microbiome DNA Enrichment Kit [1] | CpG-methylated host DNA removal | Post-extraction method; preserves microbial DNA | |
| Nucleic Acid Quantification | Qubit Fluorometric Systems [16] | Accurate DNA/RNA quantification | Essential for precise input normalization |
| PicoGreen dsDNA Assay [66] | High-sensitivity dsDNA detection | More accurate than UV absorbance for low concentrations | |
| Sample Processing | TIANamp Micro DNA Kit [16] | Microbial DNA extraction | Optimized for low-biomass samples |
| ZymoBIOMICS Reference Communities [1] | Process controls and spike-ins | Quantifiable standards for recovery calculations | |
| Sequencing Platforms | MGISEQ-2000 [16] | High-throughput sequencing | Compatible with various host depletion methods |
| Nanopore Technologies [16] | Real-time sequencing | Rapid turnaround for clinical applications |
Implementing robust host DNA depletion strategies is essential for advancing analytical sensitivity in chemogenomic NGS research. The methodologies and troubleshooting guides presented here provide a framework for significantly improving Limits of Detection and microbial recovery rates. By systematically addressing the fundamental challenge of host DNA background, researchers can enhance the reliability of pathogen detection in complex samples, ultimately supporting more sensitive diagnostics and accelerating drug development efforts. Regular validation using spiked controls and statistical LOD determination ensures ongoing optimization of these critical analytical parameters.
Quantitative Comparison: wcDNA vs. cfDNA Performance
The table below summarizes key performance metrics for whole-cell DNA (wcDNA) and cell-free DNA (cfDNA) approaches in clinical next-generation sequencing applications, particularly when combined with host depletion methods.
| Performance Metric | wcDNA with Host Depletion | cfDNA (Plasma) | Notes & Context |
|---|---|---|---|
| Pathogen Detection Sensitivity | 100% (8/8 sepsis samples) [5] [19] | Inconsistent sensitivity; not significantly enhanced by filtration [5] [19] | In gDNA-based mNGS for sepsis; culture-positive samples |
| Average Microbial Read Count | ~9,351 RPM [5] [19] | ~1,251-1,488 RPM [5] [19] | RPM: Reads per Million |
| Host DNA Background | Drastically reduced (>99% WBC removal) [5] [19] | Inherently lower than whole blood, but not enrichable via filtration [19] | wcDNA benefit relies on pre-extraction host depletion |
| Detection of CNVs/Amplifications | Well-detected from tumor tissue [67] | Feasible and concordant with tumor WGS [67] | Demonstrated in neuroblastoma (e.g., MYCN, CDK4) [67] |
| Detection of Somatic SNVs/Indels | Standard approach [67] | High concordance with tumor tissue; can reveal sub-clonal variants [67] | e.g., Rare MET p.R970C variant found in cfDNA but not in primary tumor WGS [67] |
| Informedness for Intracellular Pathogens | Superior for pathogens like Mycobacterium tuberculosis [16] | Less suitable | Host depletion enables lysis of host cells to release intracellular pathogen DNA [16] |
Detailed Experimental Protocols
Protocol 1: Host Depletion-Assisted wcDNA mNGS for Pulmonary Tuberculosis
This protocol, adapted from a study on bronchoalveolar lavage fluid (BALF) samples, uses saponin-based host cellular lysis to improve detection of intracellular pathogens [16].
- Sample Pre-processing:
- Add Sputasol (Oxoid) to the BALF sample.
- Incubate at room temperature for 2-5 minutes to lyse host cells [16].
- Centrifugation and Microbial Pellet Isolation:
- Centrifuge the treated sample.
- Resuspend the resulting pellet containing microbial cells [16].
- DNA Extraction:
- Lyse the microbial cells.
- Extract DNA using the TIANamp Micro DNA Kit (TIANGEN Biotech) or similar [16].
- Library Construction and Sequencing:
- Construct DNA libraries using the VAHTS Universal Plus DNA Library Prep Kit for MGI (Vazyme).
- Sequence on a platform such as the MGISEQ-2000 sequencer with a single-ended 50-cycle kit [16].
Protocol 2: ZISC-Based Filtration for wcDNA mNGS in Sepsis
This workflow utilizes a novel zwitterionic interface self-assemble coating (ZISC) filter to deplete white blood cells from whole blood, significantly enriching microbial content [5] [19].
- Host Cell Depletion:
- Transfer approximately 4 mL of fresh, anti-coagulated whole blood into a syringe securely connected to the ZISC-based fractionation filter (e.g., Devin filter from Micronbrane).
- Gently depress the plunger to push the blood sample through the filter into a collection tube. This step achieves >99% removal of white blood cells [5] [19].
- Differential Centrifugation for Microbial Enrichment:
- Centrifuge the filtered blood at low speed (e.g., 400g for 15 minutes) to isolate plasma.
- Transfer the plasma to a new tube and perform high-speed centrifugation (e.g., 16,000g) to obtain a microbial cell pellet [19].
- DNA Extraction and Library Preparation:
- Extract DNA from the pellet using a microbial DNA enrichment kit (e.g., ZISC-based Microbial DNA Enrichment Kit).
- Prepare sequencing libraries using an ultra-low input library prep kit (e.g., Ultra-Low Library Prep Kit from Micronbrane).
- Sequence on a platform such as the Illumina NovaSeq6000 or MiSeq [5] [19].
Protocol 3: cfDNA WGS for CNV Profiling in Neuroblastoma
This protocol outlines a non-invasive method for comprehensive genomic profiling of cancers like neuroblastoma using low-input cfDNA [67].
- Plasma and cfDNA Isolation:
- Centrifuge peripheral blood collected in EDTA tubes to separate plasma from cellular components.
- Extract cfDNA from the plasma using a commercial cfDNA extraction kit [67].
- Library Preparation and Whole-Genome Sequencing (WGS):
- Construct a sequencing library from the extracted cfDNA. The study used a low-input approach.
- Perform WGS on a platform such as the Illumina NovaSeq to an average coverage of 15x [67].
- Bioinformatic Analysis:
- Align sequence reads to the human reference genome.
- Use specialized algorithms to detect copy number variations (CNVs), somatic single nucleotide variants (SNVs), and structural variants (SVs) from the cfDNA data [67].
Experimental Workflow Visualization
Workflow Selection for wcDNA vs. cfDNA
The Scientist's Toolkit: Key Research Reagents & Kits
| Item Name | Function / Application | Specific Example / Benefit |
|---|---|---|
| ZISC-Based Filtration Device (e.g., Devin Filter) | Depletes host white blood cells from whole blood samples prior to DNA extraction. | >99% WBC removal; preserves microbial integrity; tenfold increase in microbial reads [5] [19]. |
| Saponin-Based Reagents (e.g., Sputasol) | Lyses host cells in samples like BALF to release intracellular pathogen DNA. | Crucial for improving detection of facultative intracellular pathogens like Mycobacterium tuberculosis [16]. |
| Ultra-Low Input Library Prep Kits | Constructs NGS libraries from limited or low-concentration DNA. | Essential for cfDNA WGS, enabling CNV and SNV profiling from low-input plasma samples [67]. |
| Microbial DNA Enrichment Kits | Extracts DNA from microbial pellets after host depletion. | Used post-filtration or differential centrifugation to isolate high-quality microbial gDNA for mNGS [19]. |
| Spike-in Control Standards (e.g., ZymoBIOMICS) | Monitors workflow efficiency and controls for potential background contamination. | Added to samples as an internal reference to validate microbial detection sensitivity [19]. |
Frequently Asked Questions & Troubleshooting
Q1: My pathogen detection sensitivity from blood samples is low, despite high sequencing depth. What is the most effective way to improve it?
- Problem: Overwhelming host DNA background is consuming your sequencing capacity, drowning out the microbial signal.
- Solution: Implement a pre-extraction host depletion method for wcDNA approaches. Techniques like ZISC-filtration or saponin-based lysis can remove >99% of host white blood cells, leading to a more than tenfold enrichment of microbial reads and significantly improving detection sensitivity [5] [16] [19].
- Checkpoint: Ensure you are using genomic DNA (gDNA) from the cell pellet rather than cfDNA from plasma if you plan to use host depletion filters, as these filters are not effective for cfDNA [19].
Q2: I work with intracellular pathogens like Mycobacterium tuberculosis. Why is wcDNA with host depletion superior to cfDNA for my samples?
- Problem: Intracellular pathogens reside within host cells, making their DNA difficult to access.
- Solution: The wcDNA approach, combined with a host depletion step that involves chemical lysis (e.g., with saponin), is specifically designed to break open the host cells. This releases the intracellular pathogen genomic DNA into the solution, making it available for extraction and subsequent sequencing. This method has been shown to significantly improve sensitivity and genome coverage for TB diagnosis compared to methods that do not lyse host cells [16].
Q3: Can I use cfDNA from plasma for comprehensive cancer genomic profiling, such as detecting copy number variations?
- Answer: Yes. WGS of cfDNA is a validated, non-invasive alternative to tumor tissue biopsy for detecting CNVs, somatic SNVs, and structural variants. Studies on neuroblastoma have shown high concordance between CNV profiles (including amplifications in MYCN, CDK4, and MDM2) derived from cfDNA and those from matched tumor tissue [67].
- Advantage: A key advantage of cfDNA is its ability to capture spatial heterogeneity, potentially revealing sub-clonal populations or variants present at metastatic sites that were not detected in the primary tumor biopsy [67].
Q4: When I process samples for pathogen detection, how can I monitor the efficiency of my workflow and rule out contamination?
- Solution: Consistently include a spike-in control in your workflow. Adding a known quantity of non-human, microbial cells (e.g., ZymoBIOMICS standards) to your sample during the initial processing step allows you to track DNA recovery and library preparation efficiency. Additionally, always process a negative control (no-template) sample alongside your clinical samples in each sequencing run to identify any background contamination from reagents or the environment [19].
In the field of chemogenomic next-generation sequencing (NGS) research, particularly for infectious disease diagnosis, the overwhelming presence of host DNA in clinical samples presents a significant analytical challenge. Host DNA can constitute over 90% of the genetic material in samples like blood, bronchoalveolar lavage fluid (BALF), and other human-derived specimens, drastically reducing the sequencing coverage of microbial pathogens and compromising detection sensitivity [3] [16]. This technical barrier has spurred the development of various host depletion methods, implemented through both commercial kits and laboratory-developed protocols, each with distinct performance characteristics, advantages, and limitations. The critical choice between these approaches directly impacts diagnostic accuracy, operational efficiency, and research outcomes in pathogen detection studies.
## Technical Performance Comparison
The selection between commercial kits and LDTs requires careful consideration of their operational and performance characteristics. The table below summarizes key comparative metrics:
Table 1: Performance Comparison of Host Depletion Methods
| Method Type | Host Depletion Efficiency | Microbial Read Enrichment | Labor Intensity | Cost Considerations | Typical Applications |
|---|---|---|---|---|---|
| Novel Filtration (ZISC-based) | >99% WBC removal [5] [19] | ~10-fold increase in microbial RPM (9351 vs. 925 RPM) [5] [19] | Low (integrated filtration) [5] | Medium (specialized device) | gDNA from whole blood (sepsis) [19] |
| Commercial Kit (QIAamp DNA Microbiome) | Variable (differential lysis) [19] | Moderate improvement [19] | Medium (multiple steps) | High (proprietary reagents) | Various sample types |
| Commercial Kit (NEBNext Microbiome DNA Enrichment) | Variable (CpG-methylated DNA removal) [19] | Moderate improvement [19] | Medium (multiple steps) | High (proprietary reagents) | Various sample types |
| LDT (Saponin-based HDA) | High human DNA reduction [16] | Up to 16-fold increased MTB genome coverage [16] | High (manual protocol) | Low (common reagents) | BALF for pulmonary TB [16] |
| No Host Depletion (Control) | 0% | Reference level | None | None | All sample types (baseline) |
## Experimental Protocols in Practice
### Protocol 1: ZISC-Based Filtration for gDNA from Blood
This protocol is designed for sepsis diagnosis from whole blood samples and leverages a novel zwitterionic interface coating for physical separation [19].
- Sample Preparation: Draw 3-13 mL of whole blood into a collection tube containing anticoagulant.
- Host Cell Depletion: Transfer the blood sample into a syringe attached to the ZISC-based fractionation filter (e.g., Devin filter). Gently depress the plunger to pass the blood through the filter into a sterile 15 mL collection tube. The filter retains >99% of white blood cells while allowing bacteria and viruses to pass through unimpeded [5] [19].
- Plasma Separation: Centrifuge the filtered blood at 400g for 15 minutes at room temperature. Carefully collect the supernatant (plasma).
- Microbial Pellet Recovery: Centrifuge the plasma at high speed (16,000g) to pellet microbial cells.
- DNA Extraction: Extract genomic DNA (gDNA) from the pellet using a microbial DNA enrichment kit (e.g., ZISC-based Microbial DNA Enrichment Kit) [19].
- Library Preparation & Sequencing: Proceed with standard mNGS library preparation (e.g., using an Ultra-Low Library Prep Kit) and sequence on a platform such as Illumina NovaSeq6000, aiming for at least 10 million reads per sample [5] [19].
### Protocol 2: Saponin-Based Host Depletion for BALF (LDT)
This laboratory-developed test (LDT) optimizes the detection of intracellular pathogens like Mycobacterium tuberculosis from BALF samples [16].
- Sample Pre-treatment: Add Sputasol (Oxoid) to the BALF sample. Incubate at room temperature for 2-5 minutes, or vortex for 10 minutes at 42°C [16].
- Centrifugation: Centrifuge the treated sample to pellet the contents.
- Cell Lysis and DNA Extraction: Resuspend the pellet and lyse the microbial cells. Extract total DNA using a standard kit (e.g., TIANamp Micro DNA Kit) [16].
- Quality Control: Quantify the DNA concentration using a fluorometer (e.g., Qubit 4.0).
- Library Construction & Sequencing: Prepare sequencing libraries using a standard kit (e.g., VAHTS Universal Plus DNA Library Prep Kit for MGI). Sequence on the chosen platform, such as MGISEQ-2000 [16].
Diagram 1: Host DNA Depletion Workflows. This diagram illustrates the key procedural differences between a commercial kit and a laboratory-developed protocol.
## Troubleshooting Guides and FAQs
### Troubleshooting Common Host Depletion Issues
Table 2: Troubleshooting Common NGS Preparation Issues
| Problem | Possible Causes | Recommended Solutions |
|---|---|---|
| Low microbial read count after depletion | Inefficient host cell removal; degradation of microbial DNA during processing; carryover of inhibitors. | Verify host cell count reduction; check DNA integrity; re-purify sample to remove contaminants like salts or phenol [21] [62]. |
| Low overall library yield | Poor input DNA quality/quantity; inaccurate quantification; suboptimal adapter ligation; aggressive size selection. | Use fluorometric quantification (e.g., Qubit) over UV absorbance; titrate adapter ratios; optimize bead-based cleanup parameters [21]. |
| High adapter-dimer formation | Imbalanced adapter-to-insert molar ratio; inefficient ligation; incomplete purification. | Titrate adapter concentration; ensure fresh ligase and buffer; optimize bead clean-up ratios to remove short fragments [21]. |
| Inconsistent results between technicians (LDTs) | Protocol deviations; pipetting errors; reagent degradation. | Use master mixes; implement detailed SOPs with critical steps highlighted; introduce technician checklists and "waste plates" to prevent accidental discarding of samples [21]. |
### Frequently Asked Questions (FAQs)
Q1: What is the primary benefit of using a commercial host depletion kit over an LDT? Commercial kits, such as the novel ZISC-based filter, offer standardized protocols, higher reproducibility, and are generally less labor-intensive. They provide high depletion efficiency (>99% WBC removal) and can lead to a tenfold enrichment of microbial reads, making them suitable for robust, clinical diagnostic settings [5] [19].
Q2: When might a laboratory-developed test (LDT) be preferable? LDTs are ideal for specific research applications where commercial solutions are unavailable or cost-prohibitive. They offer high customizability, as demonstrated by the saponin-based method for BALF, which provided a 16-fold increase in MTB genome coverage. LDTs are most successful in labs with established expertise for rigorous protocol optimization and validation [16].
Q3: How does high host DNA background affect my sequencing results? High host DNA proportion directly reduces the sequencing depth available for microbial genomes, decreasing the sensitivity of detection, especially for low-abundance species. With 90% host DNA, a significant number of species may remain undetected unless sequencing depth is substantially increased, raising costs [3].
Q4: My NGS library yield is low after host depletion. What should I check first? First, verify the quality and quantity of your input DNA using a fluorometric method (e.g., Qubit). Check for contaminants by assessing 260/230 and 260/280 ratios. Re-purify the sample if necessary and ensure that all enzymes and buffers are fresh and that purification bead ratios are correctly optimized [21] [62].
Q5: Can host depletion methods be used with cell-free DNA (cfDNA) for mNGS? Most pre-extraction host depletion methods, including filtration and saponin treatment, target intact host cells and are not effective for cfDNA workflows, which start with plasma. Studies show that cfDNA-based mNGS does not benefit significantly from these filtration-based host depletion techniques [19].
## The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents and Kits for Host DNA Depletion
| Reagent/Kit Name | Type | Primary Function | Example Application |
|---|---|---|---|
| Devin Filter (ZISC-based) | Commercial Kit | Physically depletes >99% of white blood cells via proprietary coating [5] [19]. | gDNA-based mNGS from whole blood for sepsis diagnostics [19]. |
| Sputasol | Laboratory Reagent | Digestant used in LDTs to liquefy mucus and release host cells in BALF samples [16]. | Sample pre-treatment for pulmonary tuberculosis diagnosis [16]. |
| QIAamp DNA Microbiome Kit | Commercial Kit | Depletes host DNA through differential lysis of human cells [19]. | Various sample types for microbiome analysis. |
| NEBNext Microbiome DNA Enrichment Kit | Commercial Kit | Enriches microbial DNA by binding and removing CpG-methylated host DNA [19]. | Various sample types for microbiome analysis. |
| Agencourt AMPure XP Beads | Laboratory Reagent | Magnetic beads used for post-fragmentation library cleanup and size selection to remove adapter dimers [21]. | Standard purification step in NGS library preparation. |
Diagram 2: Decision Logic for Host Depletion. This diagram outlines the strategic choice between commercial kits and LDTs, highlighting their inherent trade-offs.
The effective reduction of host DNA background is a cornerstone of successful pathogen detection in chemogenomic NGS research. The choice between commercial kits and laboratory-developed protocols is not a matter of superior performance in absolute terms, but of aligning methodological strengths with specific research or diagnostic needs. Commercial kits offer standardized, efficient, and user-friendly solutions ideal for clinical environments, whereas LDTs provide customizable and cost-effective alternatives for specialized research applications. By understanding the performance metrics, operational workflows, and potential pitfalls of each approach, researchers and clinicians can make informed decisions that maximize diagnostic yield and advance the field of infectious disease diagnostics.
Impact on Microbial Community Representation and Diversity Metrics
Frequently Asked Questions (FAQs)
FAQ 1: How does host DNA background affect microbial community analysis in mNGS? In samples like blood or bronchoalveolar lavage fluid (BALF), host DNA can constitute over 99% of the sequenced nucleic acids, drastically overshadowing microbial signals. This high background leads to low microbial read counts, reduced sensitivity for detecting pathogens, and wasted sequencing resources. Effective host DNA depletion is therefore critical for obtaining a true representation of the microbial community [28] [23].
FAQ 2: Can host depletion methods bias microbial diversity metrics? Yes, different host depletion methods can introduce specific taxonomic biases. For instance, some methods may significantly diminish the recovery of certain commensals and pathogens, such as Prevotella spp. and Mycoplasma pneumoniae. The choice of method can consequently alter the calculated alpha diversity metrics, such as richness and evenness, leading to an skewed representation of the original microbial community structure [23].
FAQ 3: What are the main sources of contamination in low-biomass mNGS studies? A major source of contamination is microbial DNA present in DNA extraction reagents and kits, often referred to as the "kitome." The contamination profile can vary significantly between different reagent brands and even between different manufacturing lots of the same brand. It is crucial to include negative controls (extraction blanks) in every run to identify and account for these background contaminants, which is essential for avoiding false-positive results [68].
FAQ 4: How do I choose between gDNA and cfDNA for mNGS in sepsis? Genomic DNA (gDNA) from cell pellets is highly recommended when paired with a pre-extraction host depletion method. This approach has been shown to detect all expected pathogens in clinical samples, with a more than tenfold enrichment of microbial reads compared to unfiltered samples. In contrast, cell-free DNA (cfDNA) from plasma is not amenable to pre-extraction host depletion and has demonstrated inconsistent sensitivity, making it a less reliable template for robust pathogen detection [5] [19].
Troubleshooting Guides
Problem: Low Microbial Read Count After Host Depletion
Symptoms
- Inadequate increase in microbial reads per million (RPM) following host depletion.
- Persistent high percentage of host reads in final sequencing data.
Investigation & Resolution Flowchart
Diagnostic Steps and Solutions
Verify Depletion Method Efficiency:
- Action: Compare the performance metrics of your current method against established benchmarks.
- Solution: Refer to the performance table below. If your method's host removal or bacterial retention is suboptimal, consider switching to a more balanced method like Fase or Sase [23].
Check for Incompatible Sample Types:
- Action: Confirm that your host depletion method is appropriate for your sample type.
- Solution: Pre-extraction methods (e.g., filtration, saponin lysis) are not effective for samples rich in cell-free microbial DNA, such as plasma. For cfDNA, alternative strategies are required [23].
Control for Background Contamination:
- Action: Analyze your negative control (extraction blank) samples.
- Solution: If contamination from reagents ("kitome") is high, it can mask true microbial signals. Use bioinformatics tools like Decontam to statistically identify and remove contaminant sequences from your dataset [68].
Problem: Skewed Microbial Diversity Metrics
Symptoms
- Unexpected changes in alpha diversity metrics (e.g., richness, evenness) after host depletion.
- Loss of specific microbial taxa known to be present.
Investigation & Resolution Flowchart
Diagnostic Steps and Solutions
Understand Metric Sensitivity:
- Action: Determine which specific metrics are skewed and understand what they measure.
- Solution: Richness metrics (e.g., Chao1) are highly sensitive to the total number of observed species (ASVs). Dominance and evenness metrics (e.g., Berger-Parker, Simpson) are more affected by the abundance distribution. A method that loses rare species will disproportionately affect richness estimates [69].
Identify Method-Specific Taxonomic Bias:
- Action: Consult literature on the taxonomic biases of your chosen host depletion method.
- Solution: If your study focuses on taxa known to be affected by a specific method (e.g., Prevotella spp. are diminished by S_ase), select an alternative method with a more balanced performance profile for those organisms [23].
Use a Comprehensive Set of Metrics:
- Action: Avoid relying on a single alpha diversity metric.
- Solution: Use a comprehensive suite of metrics that collectively characterize richness, phylogenetic diversity (e.g., Faith PD), dominance, and evenness. This provides a more holistic and robust view of the microbial community, reducing the risk of misinterpretation due to the limitation of any single metric [69].
Performance Data of Host Depletion Methods
Table 1: Performance of Host Depletion Methods in Respiratory Samples (BALF). Data adapted from a benchmarking study evaluating seven pre-extraction methods [23].
| Method Name | Method Description | Host DNA Removal Efficiency | Microbial Read Increase (Fold vs. Raw) | Key Taxonomic Biases / Notes |
|---|---|---|---|---|
| K_zym | HostZERO Microbial DNA Kit | Highest (99.99% / 0.9‱ remaining) | 100.3x | Best for increasing microbial reads. |
| S_ase | Saponin Lysis + Nuclease | Very High (99.99% / 1.1‱ remaining) | 55.8x | Diminishes Prevotella spp. and M. pneumoniae. |
| F_ase | 10μm Filtration + Nuclease | Not Specified | 65.6x | Most balanced performance overall. |
| K_qia | QIAamp DNA Microbiome Kit | Not Specified | 55.3x | High bacterial retention rate in OP samples. |
| O_ase | Osmotic Lysis + Nuclease | Not Specified | 25.4x | Moderate performance. |
| R_ase | Nuclease Digestion | Not Specified | 16.2x | Highest bacterial retention rate in BALF (31%). |
| O_pma | Osmotic Lysis + PMA | Not Specified | 2.5x | Least effective. |
Table 2: Impact of a Novel ZISC-Based Filtration on mNGS of Blood Samples for Sepsis Diagnosis [5] [19].
| Sample Processing Method | Average Microbial Read Count (RPM) | Pathogen Detection Rate (Culture-Positive Samples) |
|---|---|---|
| gDNA with Novel ZISC Filtration | 9,351 RPM | 100% (8/8) |
| gDNA without Filtration | 925 RPM | Not Specified |
| cfDNA with Filtration | 1,251 - 1,488 RPM | Inconsistent Sensitivity |
Research Reagent Solutions
Table 3: Essential Reagents and Kits for Host DNA Depletion in mNGS Workflows
| Product / Technology | Function | Key Application Notes |
|---|---|---|
| ZISC-Based Filtration (Devin) | Pre-extraction physical removal of host WBCs (>99%) while allowing microbes to pass. | Ideal for whole blood samples; enables gDNA-based mNGS with >10x microbial read enrichment [5] [19]. |
| MolYsis Kits (e.g., Basic5, Complete5) | Pre-extraction chemical lysis of host cells and degradation of host DNA. | Suitable for various liquid samples; a frequently mentioned standard in clinical mNGS workflows [28]. |
| QIAamp DNA Microbiome Kit | Pre-extraction differential lysis of human cells. | Compared against other methods; shows variable performance across sample types [5] [23]. |
| NEBNext Microbiome DNA Enrichment Kit | Post-extraction depletion of methylated host DNA. | Reported to have poor performance in removing host DNA from respiratory samples [23]. |
| ZymoBIOMICS Spike-in Controls | Internal positive control for DNA extraction and sequencing. | Monitors extraction efficiency and identifies technical biases; crucial for quality control [68] [19]. |
| Decontam (Bioinformatics Tool) | Computational identification and removal of contaminant sequences. | Uses statistical classification to subtract background "kitome" found in negative controls [68]. |
How is clinical performance validated for host DNA depletion methods?
Clinical validation of host DNA depletion (HDD) methods involves a direct comparison of the new metagenomic next-generation sequencing (mNGS) workflow against established diagnostic standards like culture and PCR. This process requires testing well-characterized clinical samples using both the novel HDD-mNGS method and the reference standards. The results are then compared to calculate key performance metrics, including sensitivity, specificity, and accuracy [16].
For example, in a study on pulmonary tuberculosis (PTB) diagnosis, researchers collected 105 bronchoalveolar lavage fluid (BALF) samples from suspected patients. Each sample was tested using:
- Reference Standards: Mycobacterial culture and the GeneXpert MTB/RIF (PCR-based) assay.
- Experimental Methods: Conventional mNGS, host DNA depletion-assisted mNGS (HDA-mNGS), and HDA-Nanopore sequencing.
The final clinical diagnosis, established by physicians using guidelines and all available evidence, served as the benchmark to evaluate all testing methods [16].
What quantitative improvements can be expected from effective host depletion?
Effective host DNA depletion significantly enhances key sequencing metrics, leading to better pathogen detection. The table below summarizes the performance gains observed in recent clinical studies.
Table 1: Quantitative Improvements from Host DNA Depletion in Clinical Studies
| Performance Metric | Conventional mNGS (No HDD) | With Host DNA Depletion | Clinical Sample Type | Study |
|---|---|---|---|---|
| Host Read Reduction | Baseline | >99% white blood cell removal [1] | Whole Blood (Sepsis) | [1] |
| Microbial Read Enrichment | 925 RPM [1] | 9,351 RPM (10-fold increase) [1] | Whole Blood (Sepsis) | [1] |
| Diagnostic Sensitivity | 51.2% [16] | 72.0% [16] | BALF (Tuberculosis) | [16] |
| Diagnostic Accuracy | 58.2% [16] | 74.5% [16] | BALF (Tuberculosis) | [16] |
| Pathogen Genome Coverage | Baseline | Up to 16-fold increase [16] | BALF (Tuberculosis) | [16] |
| SARS-CoV-2 Detection Rate | Not Reported | 92.9% (for Ct ≤ 35) [70] | Swab (COVID-19) | [70] |
How does HDD-mNGS performance compare to PCR and culture?
HDD-mNGS does not necessarily replace but rather complements existing methods. Its key advantage is unbiased detection, which is particularly valuable for difficult-to-culture pathogens or when previous testing is negative.
- Compared to Culture: Culture is the gold standard for many bacterial infections but is slow and has low sensitivity for fastidious organisms. HDD-mNGS can detect pathogens that are difficult or impossible to culture. In the PTB study, HDA-mNGS showed significantly higher sensitivity than culture [16].
- Compared to PCR: Multiplexed PCR panels are excellent for detecting a predefined set of pathogens. In contrast, HDD-mNGS is untargeted and can identify unexpected, novel, or rare pathogens. A study on COVID-19 showed that host-DNA removed mNGS had a detection rate of 92.9% for samples with a Ct value ≤ 35, performing comparably to approved RT-qPCR kits [70] [7]. Furthermore, mNGS can provide genomic information for strain tracing and antimicrobial resistance analysis, going beyond simple detection [16].
What are common reasons for discordant results between HDD-mNGS and culture?
Discordant results between HDD-mNGS and traditional methods are common and can arise from several technical and biological factors. The following diagram illustrates the workflow differences that lead to these discrepancies.
The most frequent scenario is a positive HDD-mNGS result with a negative culture. This is often clinically informative, not a false positive, and can be caused by:
- Prior Antibiotic Administration: Treatment before sample collection kills the pathogen, making it non-viable for culture, but its nucleic acids remain detectable by mNGS [1] [16].
- Fastidious or Intracellular Pathogens: Some bacteria, like Mycobacterium tuberculosis, are difficult to lyse or have slow growth, making them hard to culture. HDD methods designed to lyse host cells can release microbial DNA, improving mNGS detection [16].
- Non-Viable Organisms: The presence of dead pathogens can be detected by mNGS.
A negative HDD-mNGS result with a positive culture is less common but can occur due to:
- Extremely Low Pathogen Biomass: The quantity of microbial DNA is below the detection limit of the sequencing workflow, even after host depletion [16].
- Inefficient Host Depletion: If host DNA is not sufficiently removed, the sequencing depth for microbial reads remains too low [1] [33].
- Inhibition during Library Preparation: Residual contaminants in the sample can inhibit enzymes used in library preparation, leading to failure [21].
My HDD-mNGS workflow shows low microbial read counts despite high sequencing depth. How can I troubleshoot this?
Low microbial read counts after a high-depth run indicate that host depletion was inefficient. Systematically check the following areas in your protocol.
Table 2: Troubleshooting Guide for Low Microbial Reads in HDD-mNGS
| Problem Area | Potential Root Cause | Corrective Action |
|---|---|---|
| Sample Input & Quality | Sample storage conditions degraded host cells, releasing DNA [33]. | Optimize sample processing delays; use fresh samples whenever possible [1]. |
| Host Depletion Step | Inefficient lysis of host cells or incomplete DNA digestion/removal [33] [16]. | For filtration: Verify pore size and filter integrity [1]. For enzymatic methods (e.g., saponin): Titrate concentration and incubation time [16]. Include a pre-filtration step to remove free host DNA [33]. |
| DNA Extraction & Library Prep | Carryover of inhibitors (e.g., salts, phenol) from the HDD step [21]. | Perform additional clean-up steps post-extraction. Use fluorometric quantification (e.g., Qubit) over absorbance (NanoDrop) to accurately measure amplifiable DNA [21]. |
| Bioinformatics | Inaccurate read classification or use of an incomplete host reference genome [33]. | Verify the integrity and version of the host reference genome (e.g., GRCh38). Use established tools like Bowtie2 or BWA for host read alignment and removal [70] [16]. |
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents and Kits for Host DNA Depletion Workflows
| Item Name | Function / Principle | Applicable Sample Types |
|---|---|---|
| ZISC-based Filtration Device | A filter with a zwitterionic coating that selectively binds and retains host leukocytes (>99% removal) while allowing bacteria and viruses to pass through [1]. | Whole blood, other body fluids [1]. |
| Saponin | A chemical reagent that disrupts cholesterol in host cell membranes, lysing them and releasing intracellular microbes for subsequent separation [16]. | BALF, sputum, tissue samples [16]. |
| DNase I Enzyme | Degrades free host DNA fragments after host cells are lysed, while intact microbial cells are protected by their cell walls [33]. | Samples with high levels of free DNA (e.g., tissues, plasma) [33]. |
| QIAamp DNA Microbiome Kit | A commercial kit that uses differential lysis to selectively rupture human cells, followed by enzymatic degradation of the released DNA [1]. | Various sample types with high host content [1]. |
| NEBNext Microbiome DNA Enrichment Kit | Uses a methyl-CpG binding domain to bind and immobilize highly methylated host DNA, allowing unmethylated microbial DNA to be purified [1]. | Samples where microbial DNA has low methylation levels [1]. |
Multi-center Studies and Standardization Efforts Across Platforms
A significant challenge in chemogenomic Next-Generation Sequencing (NGS) research, particularly when using human blood samples, is the overwhelming abundance of host DNA. This background human DNA can consume over 95% of sequencing reads, drastically reducing the sensitivity for detecting pathogenic microbial signals and compromising data quality and research outcomes. This technical support center is designed to help researchers overcome these hurdles through standardized, evidence-based protocols and troubleshooting guides focused on effective host depletion techniques.
FAQs and Troubleshooting Guides
FAQ: Core Concepts
Q1: Why is reducing host DNA background critical for blood-based chemogenomic NGS studies? Excessive host DNA in a sample sequesters sequencing capacity, leading to poor analytical sensitivity. In a recent study, unfiltered blood samples yielded an average of only 925 microbial reads per million (RPM), while samples processed with a novel host depletion filter achieved over 10,000 microbial RPM—a tenfold enrichment that is often the difference between a conclusive result and a false negative [1].
Q2: What are the main categories of host depletion methods? Methods can be broadly classified as either pre-sequencing (physical separation or biochemical lysis) or post-sequencing (bioinformatic subtraction). Pre-sequencing methods, such as filtration, aim to remove host cells physically before DNA extraction, thereby preserving sequencing resources for microbial detection [71].
Q3: How does the performance of host depletion methods compare across different sequencing platforms? While the core biochemistry of host depletion is platform-agnostic, the efficiency of the method directly impacts the required sequencing depth. Methods that achieve higher host depletion allow for lower sequencing depths on platforms like Illumina's NovaSeq6000 or Oxford Nanopore's MinION to achieve the same diagnostic sensitivity, making projects more cost-effective [1].
Q4: What are the key quality control metrics to monitor after host depletion? Critical QC metrics include:
- Host DNA Depletion Efficiency: Measured by the percentage reduction in human DNA reads, ideally >99% white blood cell (WBC) removal [1].
- Microbial Read Retention: The number of microbial RPM post-depletion should show significant enrichment compared to an untreated control [1].
- Microbial Composition Integrity: The host depletion process must not alter the relative abundance of microbes in the sample, ensuring accurate pathogen profiling [1].
Troubleshooting Common Experimental Issues
Problem: Low Final Library Yield After Host Depletion
| Symptom | Potential Root Cause | Corrective Action |
|---|---|---|
| Low yield on Qubit/BioAnalyzer | Overly aggressive purification or size selection post-depletion. | Re-optimize bead-based cleanup ratios (e.g., adjust AMPure XP bead-to-sample ratio) and avoid over-drying the bead pellet [21]. |
| Broad or faint electropherogram peaks | Carryover of contaminants (e.g., salts, guanidine) from depletion kit reagents inhibiting enzymes. | Re-purify the DNA post-depletion using clean columns/beads with fresh wash buffers. Ensure 260/230 ratios are >1.8 [21]. |
| Low yield despite good input | Inaccurate quantification of DNA post-depletion due to contaminants. | Use fluorometric quantification (Qubit) instead of UV absorbance (NanoDrop) for accurate measurement of usable material [21] [72]. |
Problem: High Host DNA Background Persists After Depletion
| Symptom | Potential Root Cause | Corrective Action |
|---|---|---|
| High percentage of human reads | Inefficient host cell removal by the depletion method. | Verify the depletion protocol (e.g., for filtration, check flow rate, filter integrity, and blood volume capacity). Consider methods demonstrating >99% WBC removal [1]. |
| Inconsistent host depletion | Protocol deviations or human error during manual prep. | Introduce detailed SOPs with highlighted critical steps, use master mixes to reduce pipetting errors, and implement technician checklists [21]. |
| High host background in cfDNA | Using plasma cfDNA, which is not amenable to pre-extraction host-cell depletion. | Switch to a gDNA-based workflow from cell pellets, which allows for physical host cell depletion prior to DNA extraction [1]. |
Problem: Poor or Inconsistent Pathogen Detection
| Symptom | Potential Root Cause | Corrective Action |
|---|---|---|
| "No signal" or "weak signal" for spiked controls | Inhibition of enzymatic steps (ligation, PCR) by sample or reagent contaminants. | Re-purify the input sample. Ensure the DNA is eluted in water or Tris, not TE buffer, as EDTA can inhibit enzymes [72]. |
| High read count but no pathogen identified | Sporadic contamination from reagents or environment during processing. | Include negative controls (e.g., water) in every run to identify contaminating organisms, which can then be flagged and subtracted bioinformatically [73]. |
| Drop-off in sequencing read quality | Loss of microbial gDNA during multi-step depletion protocol. | Validate that the host depletion method preserves microbial integrity. Check bacterial passage efficiency through filters with plate enumeration techniques [1]. |
Experimental Protocols for Host DNA Reduction
Detailed Methodology: ZISC-Based Filtration for Host Depletion
This protocol details the use of a Zwitterionic Interface Ultra-Self-assemble Coating (ZISC)-based filtration device for depleting white blood cells from whole blood to enrich for microbial gDNA, as validated in a recent sepsis study [1].
1. Principle The novel filter coating selectively binds and retains host leukocytes and other nucleated cells without clogging, allowing bacteria and viruses to pass through unimpeded. This pre-extraction physical separation achieves >99% removal of white blood cells, significantly reducing the host DNA background [1].
2. Materials and Equipment
- Novel ZISC-based fractionation filter (e.g., Devin from Micronbrane)
- Fresh whole blood sample (3-13 mL volumes validated)
- Syringe (for connecting to the filter)
- 15 mL Falcon tube
- Low-speed and high-speed centrifuges
- Complete blood cell count analyzer (for efficiency validation)
- ZISC-based Microbial DNA Enrichment Kit or standard DNA extraction kit
3. Step-by-Step Procedure Step 1: Filtration. Transfer approximately 4 mL of whole blood into a syringe securely connected to the ZISC-based filter. Gently depress the syringe plunger to push the blood sample through the filter into a 15 mL Falcon tube [1].
Step 2: Plasma and Pellet Separation. Subject the filtered blood to low-speed centrifugation (400g for 15 minutes at room temperature) to isolate the plasma. Transfer the plasma to a new tube [1].
Step 3: Microbial DNA Extraction. Process the plasma further by high-speed centrifugation (16,000g) to obtain a sample pellet. Extract DNA from this pellet using the ZISC-based Microbial DNA Enrichment Kit or a similar validated DNA extraction method, following the manufacturer's instructions [1].
4. Performance Validation
- Host Depletion Efficiency: Measure WBC counts in pre-filtration (input) and post-filtration (output) samples using a complete blood cell count analyzer. Efficiency should consistently exceed 99% [1].
- Microbial Recovery: Use blood samples spiked with known concentrations of control bacteria (e.g., E. coli, S. aureus) or viruses. Determine bacterial counts in the filtrate via standard plate-enumeration techniques and viral concentrations via qPCR to confirm unimpeded passage [1].
Comparative Methodologies
The table below summarizes key host depletion methods based on a recent review [71].
| Method | Working Principle | Relative Efficiency | Key Limitations |
|---|---|---|---|
| ZISC-based Filtration [1] | Physical retention of host cells via a specialized zwitterionic coating. | >99% WBC removal; high microbial read preservation. | Requires specific filter device. |
| Differential Lysis (e.g., QIAamp DNA Microbiome Kit) | Selective lysis of human cells followed by degradation of released DNA. | Moderate; can be labor-intensive. | May co-lyse some gram-positive bacteria; potential for microbial DNA loss. |
| Methylated DNA Depletion (e.g., NEBNext Microbiome DNA Enrichment Kit) | Post-extraction immunoprecipitation of CpG-methylated host DNA. | Moderate reduction in host reads. | Does not reduce background from unmethylated microbial-like DNA; adds cost and step. |
| Cell-Free DNA (cfDNA) Sequencing [1] | Sequencing of non-cellular DNA from plasma, bypassing cellular background. | N/A (avoids cellular DNA). | Inconsistent sensitivity; not amenable to pre-extraction host depletion. |
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Host Depletion Workflow |
|---|---|
| ZISC-based Filtration Device [1] | The core component for physically depleting >99% of host white blood cells from whole blood samples. |
| ZISC-based Microbial DNA Enrichment Kit [1] | Optimized for DNA extraction from the microbial pellet obtained after filtration and centrifugation. |
| QIAamp DNA Microbiome Kit [1] | Provides an alternative, biochemistry-based method for host DNA removal through differential lysis. |
| NEBNext Microbiome DNA Enrichment Kit [1] | A post-extraction method that enriches for microbial DNA by removing methylated host DNA. |
| Ultra-Low Input Library Prep Kit [1] | Essential for preparing high-quality NGS libraries from the often low-yield DNA post-host-depletion. |
| ZymoBIOMICS Spike-in Control [1] | An internal reference control containing known, extremophile bacteria added to samples to monitor microbial recovery and detect inhibition. |
| AMPure XP Beads [21] | Used for post-library preparation cleanup to remove adapter dimers and select for the desired fragment size, crucial after low-input workflows. |
Workflow and Pathway Visualizations
Host DNA Depletion and mNGS Workflow
Troubleshooting High Host DNA Background
Conclusion
Effective host DNA depletion is no longer optional but essential for maximizing the diagnostic potential of metagenomic NGS in clinical and research settings. The evidence demonstrates that integrated approaches combining novel filtration technologies like ZISC-based systems with optimized bioinformatics pipelines can achieve >99% host cell removal and tenfold enrichment of microbial reads. Method selection must be guided by sample type, with wcDNA-based approaches showing superior sensitivity for bloodstream infections while enzymatic methods better preserve DNA integrity for long-read sequencing. Future directions should focus on standardizing depletion protocols, developing rapid point-of-care compatible methods, and creating universal quality metrics. As host depletion technologies continue to evolve, they will undoubtedly expand the clinical utility of mNGS for rapid pathogen identification, antimicrobial resistance profiling, and outbreak investigation, ultimately transforming our approach to infectious disease diagnosis and management.
References
- 1. Optimization of Metagenomic Next-Generation Sequencing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12436562/]
- 2. Host DNA depletion on frozen human respiratory samples ... [https://www.nature.com/articles/s42003-024-07290-3]
- 3. Impact of Host DNA and Sequencing Depth on the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6581681/]
- 4. Impact of Host DNA and Sequencing Depth on the ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2019.01277/full]
- 5. Optimization of Metagenomic Next-Generation Sequencing ... [https://pubmed.ncbi.nlm.nih.gov/40670871/]
- 6. Seven tips for avoiding DNA contamination in your PCR [https://www.takarabio.com/about/bioview-blog/tips-and-troubleshooting/avoid-dna-contamination-in-pcr?srsltid=AfmBOooCJ4I1do3pQZLJ0f7I8Hel_DC1ZiUFQqTG2yJjJvHt1yO313tg]
- 7. Clinical characteristics of the host DNA-removed ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.1016440/full]
- 8. Contaminant DNA in bacterial sequencing experiments is a ... [https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-020-0748-z]
- 9. Direct real-time quantitative PCR for measurement of host ... [https://pubmed.ncbi.nlm.nih.gov/25151232/]
- 10. Quantifying DNA? Here are Five DNA Quantification ... [https://blog.addgene.org/five-methods-for-quantifying-dna]
- 11. 5 Different DNA Quantification Methods to Consider [https://www.denovix.com/5-different-dna-quantification-methods-to-consider/]
- 12. Residual Host Cell DNA Quantitation [https://www.thermofisher.com/us/en/home/life-science/bioproduction/contaminant-and-impurity-testing/host-cell-residual-dna-quantitation.html]
- 13. Efficient and robust RNA-seq process for cultured bacteria and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3439974/]
- 14. Improving saliva shotgun metagenomics by chemical host ... [https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0426-3]
- 15. Metagenomic Sequencing for Microbial DNA in Human ... [https://www.mdpi.com/1422-0067/23/4/2181]
- 16. Host DNA depletion assisted metagenomic sequencing of ... [https://ann-clinmicrob.biomedcentral.com/articles/10.1186/s12941-025-00782-y]
- 17. Human DNA hijacking microbiota surveys: causes and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12455517/]
- 18. Host-DNA Depletion Solutions for Metagenomic Diagnostics [https://www.arcticzymes.com/application/host-cell-dna-depletion-prior-to-diagnostic-metagenomic-analysis]
- 19. Optimization of Metagenomic Next-Generation Sequencing Workflow... [https://link.springer.com/article/10.1007/s40291-025-00797-3]
- 20. Comparative analysis of metagenomic next-generation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11934473/]
- 21. How to Troubleshoot Sequencing Preparation Errors (NGS ... [https://www.cd-genomics.com/resource-sequencing-preparation-troubleshooting.html]
- 22. How-to: NGS Quality Control [https://frontlinegenomics.com/how-to-ngs-quality-control/]
- 23. Benefits and challenges of host depletion methods in ... [https://www.nature.com/articles/s41522-025-00762-2]
- 24. Common Centrifuge Issues and How to Fix Them [https://newlifescientific.com/blogs/new-life-scientific-blog/centrifuge-troubleshooting?srsltid=AfmBOop34XdK6V9h8S4dbdnEJJ_xyvnBU17WsZ7Sr52_-f5hAuQ0oNuF]
- 25. Effects of different centrifugation conditions on clinical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3123263/]
- 26. Troubleshooting Guide for Genomic DNA Extraction & ... [https://www.neb.com/en-us/tools-and-resources/troubleshooting-guides/troubleshooting-guide-for-t3010?srsltid=AfmBOorUsFwM2E_hHAAc3bWV6hVatcTdcl7M9OjdF4fJAQuNkps6uKM2]
- 27. Restriction enzyme digestion of host DNA enhances universal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6142370/]
- 28. Key Insights from ESCMID 2025: Advancing Metagenomic ... [https://www.vhbio.com/escmid-2025-advancing-metagenomic-sequencing-clinical-practice/]
- 29. NGS Library Preparation Support—Troubleshooting [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/next-generation-sequencing-support-center/ngs-library-preparation-support/ngs-library-preparation-support-troubleshooting.html]
- 30. Mechanical vs. enzymatic fragmentation approaches [https://www.idtdna.com/page/support-and-education/decoded-plus/mechanical-vs-enzymatic-fragmentation-approaches-which-method-best-improves-library-quality/]
- 31. Technology [https://puriblood.com/en/about/technology/]
- 32. Bioinformatic approaches to blood and tissue microbiome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12018304/]
- 33. Metagenomic Sequencing: Strategies for Host DNA Removal [https://www.cd-genomics.com/microbioseq/resource-metagenomic-sequencing-host-dna-removal.html]
- 34. Targeted decontamination of sequencing data with CLEAN [https://pmc.ncbi.nlm.nih.gov/articles/PMC12288876/]
- 35. Contamination Issue in Viral Metagenomics [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2021.745076/full]
- 36. Guidelines for preventing and reporting contamination in ... [https://www.nature.com/articles/s41564-025-02035-2]
- 37. Comparative Study of Host Depletion DNA for Microbial... Methods [https://www.researchsquare.com/article/rs-5991450/v1]
- 38. Metagenomic next-generation sequencing reveals ... [https://www.nature.com/articles/s41598-025-21722-y]
- 39. Advantages and challenges of metagenomic sequencing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9527156/]
- 40. Clinical impact of metagenomic next-generation ... [https://www.sciencedirect.com/science/article/pii/S2405844024113187]
- 41. High-Molecular-Weight DNA : Extraction ... - CD Genomics Optimized [https://www.cd-genomics.com/resource-high-molecular-weight-dna-extraction.html]
- 42. Protocol for Optimized in Three Theobroma Species DNA Extraction [https://en.bio-protocol.org/en/bpdetail?id=5297&type=0]
- 43. High- yield protocol for Azadirachta... - Academia.edu DNA extraction [https://www.academia.edu/3064-9765/2/2/10.20935/AcadMolBioGen7730]
- 44. Maximizing DNA for Biobanking Applications | Genetics And... Yield [https://www.labroots.com/trending/genetics-and-genomics/27179/maximizing-dna-yield-biobanking-applications]
- 45. and comparison of genomic Optimization fro... DNA extraction [https://www.degruyterbrill.com/document/doi/10.1515/cclm-2025-1079/html]
- 46. Benefits and challenges of host depletion methods in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12234985/]
- 47. Host DNA depletion efficiency of microbiome ... [https://www.sciencedirect.com/science/article/abs/pii/S0167701219310693]
- 48. Troubleshooting Guide for Genomic DNA Extraction & ... [https://www.neb.com/en-us/tools-and-resources/troubleshooting-guides/troubleshooting-guide-for-t3010?srsltid=AfmBOooA5kZTUIfJ1GJif4Z0cVursa8Gg4Og5q2PZIqC4dq-1H35no_n]
- 49. Troubleshooting Low DNA Yield from Blood Samples [https://www.cambrianbioworks.com/blog/troubleshooting-low-dna-yield-from-blood-samples-a-guide-for-clinical-labs]
- 50. Sample Preparation for NGS – A Comprehensive Guide [https://frontlinegenomics.com/sample-preparation-for-ngs-a-comprehensive-guide/]
- 51. Mastering DNA chromatogram analysis in Sanger ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10643650/]
- 52. High Sensitivity of Shotgun Metagenomic Sequencing in ... [https://www.sciencedirect.com/science/article/pii/S167202292200119X]
- 53. Systematic Assessment of Matrix Effect, Recovery, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12613129/]
- 54. Incomplete human reference genomes can drive false sex ... [https://www.nature.com/articles/s41467-025-56077-5]
- 55. De-biasing microbiome sequencing data: bacterial ... [https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-024-01998-4]
- 56. Development of reference-based model for improved ... [https://www.sciencedirect.com/science/article/pii/S0963996925007185]
- 57. FAQs Next Generation Sequencing [https://eurofinsgenomics.eu/en/eurofins-genomics/product-faqs/next-generation-sequencing/inview-metagenome/host-contamination/]
- 58. PCR Troubleshooting Guide [https://www.neb.com/en-us/tools-and-resources/troubleshooting-guides/pcr-troubleshooting-guide?srsltid=AfmBOor0LCBi1dLE8RF3iuzqIThQU9qMl4TZNIXpql_wYL_RWqH2BuTt]
- 59. Contamination in Sequence Databases - NCBI - NIH [https://www.ncbi.nlm.nih.gov/tools/vecscreen/contam]
- 60. Metagenomic Next-Generation Sequencing in Infectious Diseases: Clinical Applications, Translational Challenges, and Future Directions [https://www.mdpi.com/2075-4418/15/16/1991]
- 61. Avoid Costly Errors in NGS Sample Handling and Library ... [https://seqwell.com/avoid-costly-errors-in-ngs-sample-handling-and-library-prep/]
- 62. What's wrong with my NGS? [https://www.ogt.com/resources/ngs-resources-and-support/ngs-support/whats-wrong-with-my-ngs/]
- 63. Integrating Artificial Intelligence in Next-Generation Sequencing [https://pmc.ncbi.nlm.nih.gov/articles/PMC12191491/]
- 64. DNA Purification | DNA Extraction Methods [https://www.promega.com/resources/guides/nucleic-acid-analysis/dna-purification/]
- 65. Method Detection Limit - Frequent Questions [https://www.epa.gov/cwa-methods/method-detection-limit-frequent-questions]
- 66. Microplate Assays for Nucleic Acids [https://www.thermofisher.com/us/en/home/life-science/cell-analysis/fluorescence-microplate-assays/microplate-assays-nucleic-acids.html]
- 67. Comparative analysis of whole-genome sequencing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12081241/]
- 68. Deciphering the impact of contaminating microbiota in DNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12502690/]
- 69. Key features and guidelines for the application of microbial ... [https://www.nature.com/articles/s41598-024-77864-y]
- 70. Clinical characteristics of the host DNA-removed ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9705594/]
- 71. Advances in Host Depletion and Pathogen Enrichment ... [https://www.sciencedirect.com/science/article/pii/S1525157824001284]
- 72. retrogen sequencing troubleshooting guide and suggestions [https://retrogen.net/resources/genomic/]
- 73. Seven-year performance of a clinical metagenomic next- ... [https://www.nature.com/articles/s41591-024-03275-1]