Precision Calling: Optimizing NGS Variant Accuracy in Chemogenomic Screens for Drug Discovery
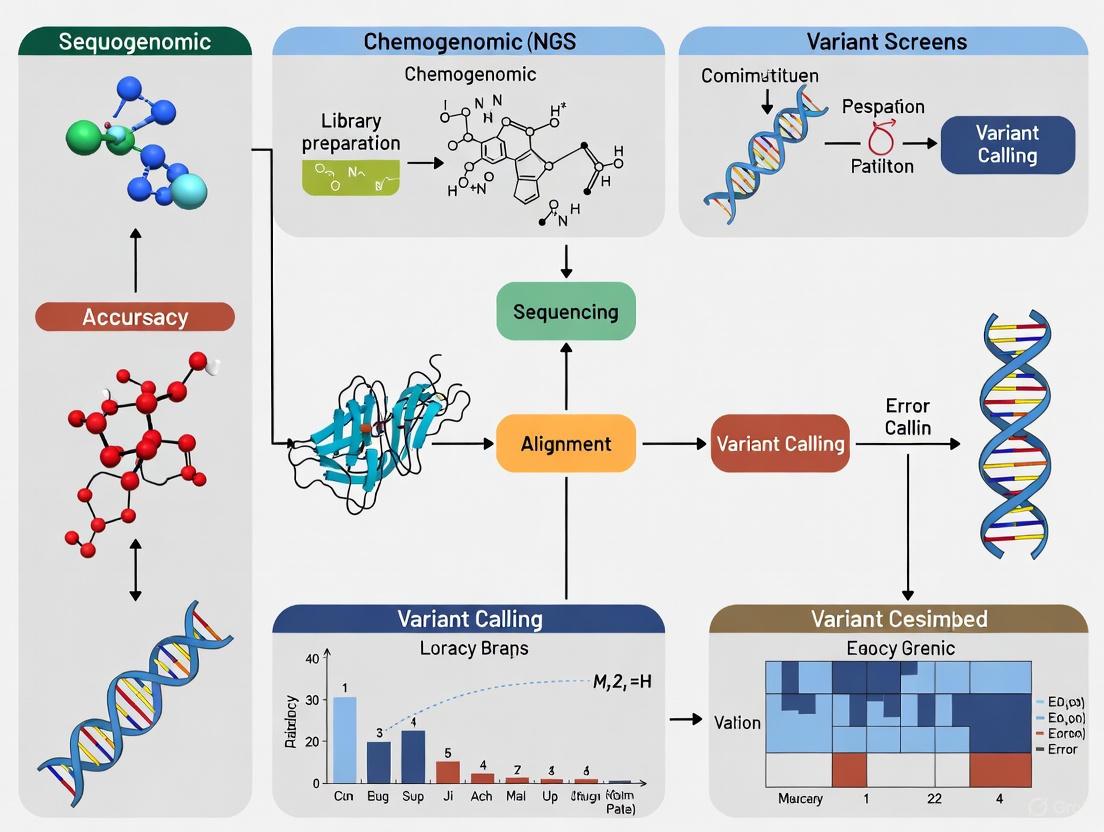
Next-generation sequencing (NGS) variant calling is foundational to interpreting chemogenomic screens, where accurately linking genetic perturbations to compound sensitivity is paramount.
Precision Calling: Optimizing NGS Variant Accuracy in Chemogenomic Screens for Drug Discovery
Abstract
Next-generation sequencing (NGS) variant calling is foundational to interpreting chemogenomic screens, where accurately linking genetic perturbations to compound sensitivity is paramount. This article provides a comprehensive guide for researchers and drug development professionals, covering the core principles of variant calling, best-practice methodologies for different variant types, strategies for troubleshooting and optimizing pipelines, and rigorous frameworks for validation and benchmarking. By synthesizing current best practices and emerging trends, including the impact of AI and multi-omics integration, this resource aims to empower scientists to enhance the reliability and actionability of their data, thereby accelerating target identification and therapeutic development.
The Bedrock of Precision: Core Concepts and Challenges in NGS Variant Calling
In chemogenomics, which explores the complex interactions between chemical compounds and biological systems, the accurate identification of genomic variants is a foundational pillar. Next-generation sequencing (NGS) has become an indispensable tool for uncovering the genetic determinants of drug response, resistance, and toxicity. However, the inherent limitations of sequencing technologies and the diverse nature of genomic alterations present significant challenges. Variants are broadly categorized by size and type: Single Nucleotide Variants (SNVs) involve changes to a single base; Insertions and Deletions (Indels) are typically under 50 bp; Copy Number Variations (CNVs) are large-scale duplications or deletions; and Structural Variations (SVs), including balanced rearrangements like inversions, are generally 50 bp or larger [1]. The sophistication of variant calling is ever-increasing, yet the performance of detection algorithms varies dramatically depending on the variant type, genomic context, and sequencing technology used [2] [1]. This guide provides an objective comparison of variant calling performance, synthesizing recent benchmarking data to inform robust experimental design in chemogenomic screens. A precise understanding of the variant landscape enables researchers to better correlate genetic features with compound activity, ultimately accelerating drug discovery and development.
Performance Comparison of Variant Callers and Technologies
The accuracy of variant detection is influenced by a complex interplay of sequencing technology and the computational algorithm employed. The tables below summarize key performance metrics from recent benchmarking studies.
Table 1: Comparative Performance of Short-Read and Long-Read Sequencing for Variant Detection
| Variant Type | Sequencing Technology | Key Performance Findings | Notable Top-Performing Tools |
|---|---|---|---|
| SNVs | Short-Read (Illumina) | High accuracy, similar to long-reads in non-repetitive regions [1]. | DeepVariant [1] [3] |
| Long-Read (ONT/PacBio) | Matches or exceeds short-read accuracy; deep learning tools achieve F1 scores >99.9% [3]. | Clair3, DeepVariant [3] | |
| Indels | Short-Read (Illumina) | Recall for insertions >10 bp is poor compared to long-reads; performance decreases in repetitive regions [1]. | GATK [1] |
| Long-Read (ONT/PacBio) | Superior accuracy for all indels; deep learning tools achieve F1 scores >99.5% [3]. | Clair3, DeepVariant [3] | |
| Structural Variants (SVs) | Short-Read (Illumina) | Significantly lower recall in repetitive regions; union of multiple algorithms enhances detection [4] [1]. | DELLY, LUMPY, Manta, GRIDSS [4] |
| Long-Read (ONT/PacBio) | Higher sensitivity and precision, especially in repetitive regions and for complex SVs [1]. | cuteSV, Sniffles, pbsv [1] |
Table 2: Performance of Specific SV Detection Algorithms on Short-Read Data (GIAB Benchmark)
| Algorithm/Strategy | Recall (DEL) | Precision (DEL) | F1 Score (DEL) | Recall (INS) | Precision (INS) | F1 Score (INS) |
|---|---|---|---|---|---|---|
| DELLY | 0.62 | 0.91 | 0.74 | 0.14 | 0.93 | 0.24 |
| LUMPY | 0.69 | 0.89 | 0.78 | 0.21 | 0.92 | 0.34 |
| Manta | 0.76 | 0.95 | 0.84 | 0.43 | 0.97 | 0.60 |
| DRAGEN (Commercial) | 0.82 | 0.97 | 0.89 | 0.60 | 0.98 | 0.75 |
| Union of 3 Algorithms | 0.86 | 0.91 | 0.88 | 0.65 | 0.92 | 0.76 |
Data adapted from Duan et al., 2025 [4]. The union strategy combined Manta, MELT, and GRIDSS. DEL=Deletion, INS=Insertion.
Key Insights from Performance Data
- The Superiority of Ensemble Methods for SVs: For SV detection on short-read data, no single algorithm is universally optimal. A union strategy combining multiple callers (e.g., Manta, MELT, and GRIDSS) can achieve performance comparable to, and sometimes surpassing, sophisticated commercial software like DRAGEN, particularly for insertions [4]. Interestingly, expanding the ensemble beyond three well-chosen algorithms does not necessarily improve performance [4].
- The Rise of Deep Learning and Long-Reads: For SNVs and Indels, deep learning-based variant callers (Clair3, DeepVariant) applied to high-accuracy long-read data (e.g., ONT super-accuracy mode) are now setting a new standard, outperforming traditional methods and even challenging the historical primacy of Illumina short-reads [3].
- Context Matters: Repetitive Regions Are a Major Challenge: A critical finding across studies is that the performance of short-read-based variant callers for indels and SVs deteriorates significantly in repetitive regions, such as segmental duplications and simple tandem repeats [1]. Long-read technologies, by spanning these repetitive elements, maintain high accuracy [1].
Experimental Protocols for Benchmarking Variant Callers
To generate the comparative data presented in this guide, benchmarking studies follow rigorous and standardized experimental workflows. The following diagram and protocol outline the common steps involved in evaluating variant detection performance.
Detailed Benchmarking Methodology
- Sample and Data Preparation: Benchmarking relies on well-characterized reference samples from consortia like the Genome in a Bottle (GIAB) Consortium (e.g., NA12878, HG002) [4] [1]. The same DNA extraction is used for both short-read (Illumina) and long-read (PacBio HiFi, ONT) sequencing to prevent culture-induced mutations from biasing results [3].
- Read Alignment and Variant Calling: Sequencing reads are aligned to a reference genome (GRCh37/hs37d5). Short reads are typically aligned with
bwa mem, while long reads are aligned withminimap2[1]. A wide array of variant calling algorithms (e.g., 6 for SNVs, 12 for indels, 13 for SVs) are then run on the aligned data [1]. - Truth Set Construction: A high-confidence set of true variants is essential for evaluation. This is created by integrating data from multiple sources, including:
- GIAB benchmark sets [1].
- Long-read-based haplotype-resolved assemblies (e.g., from HGSVC) [1].
- Variants commonly identified by multiple long-read-based callers to ensure precision [1].
- For bacterial studies, a "pseudo-real" truthset may be generated by projecting real variants from a closely related donor genome onto a gold-standard reference [3].
- Performance Evaluation and Manual Inspection: Variant calls from each tool are compared against the truth set using
vcfevalorvcfdistto calculate precision, recall, and F1 scores [3]. To ensure the highest quality, a subset of variants, particularly indels, is often validated through manual visual inspection using tools like the Integrative Genomics Viewer (IGV) [1].
Successful variant detection and annotation require a suite of computational tools and genomic resources. The following table details key solutions used in the featured experiments.
Table 3: Key Research Reagent Solutions for NGS Variant Detection
| Item Name | Function/Description | Application in Variant Detection |
|---|---|---|
| GIAB Reference Materials | Genomic DNA from well-characterized human cell lines (e.g., HG002). | Provides a gold-standard benchmark for validating variant calls across platforms and algorithms [1]. |
| Variant Call Format (VCF) | A standardized text file format for storing gene sequence variations. | The primary output of variant callers; used for interoperability between different analysis tools [5]. |
| Integrative Genomics Viewer (IGV) | A high-performance visualization tool for interactive exploration of large genomic datasets. | Enables manual inspection and validation of variant calls by visualizing read alignments [2]. |
| Ensembl VEP & ANNOVAR | Command-line tools for determining the functional consequences of variants (e.g., missense, frameshift). | Critical for annotating VCF files to predict the impact of variants on genes and proteins [6]. |
| CAVA (Clinical Annotation of VAriants) | A tool for providing standardized, clinically appropriate annotation of NGS data. | Resolves inconsistencies in indel annotation, ensuring compatibility with historical clinical data [5]. |
| BioRender | A web-based tool for creating scientific figures and illustrations. | Used to generate professional diagrams of workflows, signaling pathways, and results for publications. |
The landscape of NGS variant calling is evolving rapidly, driven by advancements in long-read sequencing and sophisticated deep learning algorithms. For chemogenomics researchers, the choice of technology and computational pipeline must be tailored to the variant types of interest. While short-read sequencing combined with ensemble calling strategies remains a powerful and cost-effective approach for population-scale studies, long-read technologies offer a compelling alternative for comprehensive variant discovery, particularly in complex genomic regions. The emerging paradigm emphasizes that there is no single "best" tool, but rather a need for refined, context-aware strategies. As the field moves forward, the integration of multiple data types and continuous benchmarking against curated truth sets will be paramount for defining the variant landscape with the accuracy required to power the next generation of chemogenomic discoveries.
In chemogenomic screens, which systematically explore the interactions between chemical compounds and genetic variants, the accuracy of next-generation sequencing (NGS) data analysis is paramount. These screens rely on precise variant calling to identify genetic modifiers of drug response, potential drug targets, and mechanisms of resistance. The standard NGS data analysis pipeline serves as the foundation for extracting meaningful biological insights from raw sequencing data, transforming billions of short DNA fragments into accurate genetic variants [7] [8]. Even minimal error rates in sequencing data—seemingly low at 0.1-1%—can translate to thousands of incorrect base calls across the human genome, severely compromising the identification of true somatic mutations or single nucleotide polymorphisms (SNPs) in chemogenomic studies [7]. This article provides a comprehensive comparison of current NGS analysis methodologies, focusing on their performance characteristics and providing supporting experimental data to guide researchers in selecting optimal pipelines for precision oncology and chemogenomics research.
The Standard NGS Analysis Pipeline: A Step-by-Step Breakdown
The journey from raw sequencing output to biological insights follows a structured pathway with distinct computational steps. Each stage addresses specific data quality challenges and prepares the data for subsequent analysis, with the overall workflow managed by specialized systems that ensure reproducibility and efficiency [9].
Quality Control and Adapter Trimming
The initial critical stage involves assessing data quality and removing technical sequences. Raw sequencing data in FASTQ format contains not only sequence reads but also quality scores for each base, and potential contaminants such as adapter sequences [9].
- Purpose: To identify low-quality bases, sequence bias, and over-represented sequences that could skew downstream analysis [9].
- Common Tools: FastQC, fastp, and MultiQC are widely employed for quality assessment [9]. FastQC provides comprehensive quality metrics including per-base sequence quality, sequence duplication levels, and adapter contamination [9].
- Adapter Trimming: Tools like Cutadapt, Trimmomatic, and fastp remove adapter sequences and trim low-quality bases, significantly improving data quality for alignment [9].
Alignment to a Reference Genome
Processed reads are then mapped to a reference genome to identify their genomic origins.
- Purpose: To determine the precise location in the genome from which each sequencing read originated [9].
- Alignment Tools: BWA-Mem, Bowtie, HISAT2, and STAR are commonly used aligners [9]. BWA-Mem is particularly prevalent for DNA sequencing data [9] [8].
- Output: The alignment process produces BAM files (Binary Alignment/Map format), which store the aligned sequences and their mapping qualities [8].
Variant Calling
This crucial step identifies genetic differences (variants) between the sequenced sample and the reference genome.
- Purpose: To detect single nucleotide variants (SNVs), insertions and deletions (indels), and other genetic variations [9] [8].
- Approaches: Variant callers employ diverse algorithms, from traditional statistical methods to emerging deep learning approaches:
- Germline Variant Calling: GATK HaplotypeCaller, FreeBayes, and Platypus are established tools [8].
- Somatic Variant Calling: Mutect2 is specifically designed for identifying cancer-associated mutations [9].
- Deep Learning Methods: Clair3 and DeepVariant use convolutional neural networks for improved accuracy [3].
Variant Annotation and Filtering
Identified variants undergo biological interpretation through annotation and filtering.
- Purpose: To predict the functional consequences of variants and filter out technical artifacts [9].
- Annotation Tools: VEP (Variant Effect Predictor), ANNOVAR, and SnpEff provide information on variant consequences, such as whether they affect protein coding, regulatory regions, or splice sites [9].
- Filtering Resources: Databases like dbSNP, 1000 Genomes, and gnomAD help distinguish common polymorphisms from potentially disease-relevant mutations [9].
Comparative Analysis of Variant Calling Technologies
Performance Benchmarking Across Platforms and Tools
Recent comprehensive benchmarking studies reveal significant differences in variant calling accuracy across platforms and computational methods. A landmark study evaluating 14 bacterial species demonstrated that deep learning-based variant callers applied to Oxford Nanopore Technologies (ONT) data outperformed traditional methods and even surpassed Illumina sequencing accuracy for both SNPs and indels [3].
Table 1: Performance Comparison of Variant Calling Methods for Oxford Nanopore Technologies Data
| Variant Caller | Type | SNP F1 Score (%) | Indel F1 Score (%) | Best For |
|---|---|---|---|---|
| Clair3 | Deep Learning | 99.99 | 99.53 | Overall accuracy |
| DeepVariant | Deep Learning | 99.99 | 99.61 | Indel calling |
| Medaka | Deep Learning | 99.80 | 98.20 | Fast processing |
| NanoCaller | Deep Learning | 99.70 | 97.50 | Complex variants |
| BCFtools | Traditional | 99.30 | 80.10 | Standard SNPs |
| FreeBayes | Traditional | 98.90 | 85.60 | Germline variants |
| Longshot | Traditional | 99.50 | 90.20 | Long read haplotyping |
Data adapted from eLife benchmarking study [3]
For Illumina platforms, the Genome Analysis Toolkit (GATK) remains a widely adopted solution, particularly known for its robust variant discovery and genotyping capabilities [10] [9]. The Broad Institute's Best Practices workflow provides a standardized approach for processing Illumina data, though evaluations suggest that the improvements from certain preprocessing steps like base quality score recalibration may be marginal considering their computational cost [8].
Impact of Sequencing Technology on Variant Calling Accuracy
Different sequencing technologies exhibit characteristic error profiles that directly impact variant calling accuracy:
- Illumina Platforms: Generally display low error rates (0.26%-0.8%) but may show substitution errors in AT-rich or CG-rich regions [7] [11].
- Ion Torrent: Similar to 454 pyrosequencing, has limitations in homopolymer regions with an error rate of approximately 1.78% [7] [11].
- SOLiD: Utilizes a two-base encoding system that achieves a lower error rate of about 0.06%, though with shorter read lengths [7].
- Oxford Nanopore: Historically had higher error rates, but with the latest R10.4.1 pore and super-accuracy basecalling, median read identities can reach 99.93% (Q32) for duplex reads, enabling F1 scores >99.5% for both SNPs and indels when using deep learning variant callers [3].
Table 2: Sequencing Platform Error Profiles and Characteristics
| Platform | Typical Error Rate | Error Type | Read Length | Best Application |
|---|---|---|---|---|
| Illumina | 0.26%-0.8% | Substitutions | Short (36-300 bp) | High-throughput screening |
| Ion Torrent | ~1.78% | Homopolymer indels | Short (200-400 bp) | Targeted sequencing |
| SOLiD | ~0.06% | Substitutions | Short (75 bp) | Variant validation |
| PacBio | Variable | Random errors | Long (10-25 kb) | Structural variants |
| Oxford Nanopore | <0.1% (sup duplex) | Random errors | Long (10-30 kb) | Comprehensive variant detection |
Data synthesized from multiple sources [7] [11] [3]
Experimental Protocols for Benchmarking Variant Calling Performance
Establishing Gold Standard Reference Sets
Rigorous benchmarking of variant calling pipelines requires well-characterized reference datasets where "ground truth" variants are known. The Genome in a Bottle (GIAB) consortium and Platinum Genomes project provide benchmark variants for human genomes, particularly for the extensively characterized NA12878 sample [8]. For bacterial genomics, a novel approach involves projecting real variants from closely related donor genomes (with ~99.5% average nucleotide identity) onto gold standard reference assemblies, creating biologically realistic variant distributions for benchmarking [3].
Protocol: Truthset Generation for Bacterial Genomes
- Generate high-quality reference assemblies using both ONT and Illumina reads
- Select donor genome with closest to 99.5% average nucleotide identity
- Identify variants between sample and donor using minimap2 and MUMmer
- Intersect variant sets and filter to remove overlaps and indels >50bp
- Apply variant truthset to sample's reference to create mutated reference
- Validate mutated reference against original donor genome [3]
Performance Assessment Metrics
Variant calling accuracy is typically evaluated using standard classification metrics:
- Precision: The proportion of called variants that are true positives (fewer false positives)
- Recall: The proportion of true variants that are detected (fewer false negatives)
- F1 Score: The harmonic mean of precision and recall, providing a balanced metric [3]
Specialized tools like vcfdist are used for variant comparison, properly handling complex variants and differences in variant representation [3].
Visualization of the NGS Data Analysis Workflow
The following diagram illustrates the complete standard NGS data analysis pipeline, from raw sequencing data to biological insights, including key decision points for tool selection:
Diagram 1: Standard NGS analysis workflow with key computational steps and representative tools for each stage.
Workflow Management Systems
Modern NGS analysis relies on workflow managers that ensure reproducibility, scalability, and efficient resource utilization:
- Nextflow: A domain-specific language that enables portable and reproducible workflows, with active community-developed pipelines available through nf-core [9].
- Snakemake: A Python-based workflow management system that excels in creating transparent and flexible analysis pipelines [9].
- Galaxy: A web-based platform that provides an accessible interface for bioinformatics analysis, particularly valuable for researchers with limited computational expertise [9].
Accurate variant interpretation depends on comprehensive biological databases:
- Reference Genomes: GRCh38 (human) and species-specific references provide the coordinate systems for alignment [9].
- Population Databases: gnomAD, 1000 Genomes, and dbSNP contain information on population allele frequencies, helping filter common polymorphisms [9] [8].
- Functional Annotation: Ensembl, RefSeq, and UniProt provide gene models and functional information for consequence prediction [9].
Table 3: Essential Bioinformatics Tools and Resources for NGS Analysis
| Category | Tool/Resource | Primary Function | Application in Chemogenomics |
|---|---|---|---|
| Workflow Management | Nextflow, Snakemake | Pipeline automation and reproducibility | Ensures consistent analysis across screens |
| Quality Control | FastQC, MultiQC | Quality assessment and reporting | Identifies batch effects and technical artifacts |
| Alignment | BWA-Mem, STAR | Read mapping to reference genome | Critical for accurate variant detection |
| Variant Calling | GATK, Clair3, DeepVariant | Genetic variant identification | Detects drug-resistance mutations |
| Variant Annotation | VEP, ANNOVAR | Functional consequence prediction | Prioritizes variants affecting drug targets |
| Visualization | IGV | Manual variant inspection | Validates candidate variants in context |
| Benchmarking | vcfdist, GIAB resources | Performance assessment | Quantifies pipeline accuracy |
The field of NGS data analysis is undergoing rapid transformation, with deep learning approaches demonstrating remarkable improvements in variant calling accuracy. The latest benchmarking evidence suggests that Oxford Nanopore sequencing combined with deep learning variant callers like Clair3 can achieve F1 scores exceeding 99.5% for both SNPs and indels, potentially surpassing the accuracy of traditional Illumina-based pipelines [3]. For chemogenomic screens, where identifying true genetic modifiers of drug response is critical, these technological advances promise enhanced sensitivity for detecting low-frequency variants and improved specificity in distinguishing genuine mutations from sequencing artifacts.
Future developments will likely focus on integrating multimodal data, improving scalability for large-scale screens, and enhancing the detection of complex structural variations. As sequencing technologies continue to evolve, the establishment of rigorous benchmarking standards and reproducible analysis pipelines will remain essential for maximizing the value of NGS data in chemogenomics research and precision oncology.
In modern drug discovery, next-generation sequencing (NGS) has become an indispensable tool for identifying and validating potential drug targets. The process of variant calling—identifying genetic variations from sequencing data—serves as the critical foundation upon which target discovery rests [12]. Inaccurate variant calling can create a cascade of errors, leading research down unproductive pathways, overlooking genuine therapeutic targets, and ultimately wasting substantial resources during development [13]. Within chemogenomic screens, where researchers systematically study interactions between chemical compounds and genetic variants, variant calling accuracy becomes particularly crucial as it directly influences our understanding of disease mechanisms and potential therapeutic interventions [12] [14].
This guide examines how variant calling errors impact drug target discovery, compares the performance of various variant calling methods, and provides evidence-based recommendations for implementing optimal practices in genomic research. By understanding the sources and consequences of these errors, researchers and drug development professionals can make informed decisions that enhance the reliability and efficiency of their target discovery pipelines.
The Critical Role of Variant Calling in Drug Discovery Pipelines
From Sequence to Therapy: How Variant Calling Informs Target Discovery
Variant calling provides the essential genetic insights that drive multiple stages of the drug discovery pipeline [12]. Initially, population-scale sequencing studies leverage electronic health records to identify associations between genetic variants and specific disease phenotypes, highlighting potential therapeutic targets [12]. Once candidate targets emerge, researchers use loss-of-function mutation detection in combination with phenotype studies to validate target relevance and predict potential effects of therapeutic inhibition [12]. During drug design, variant calling informs this process by revealing details about genome structure, genetic variations, gene expression profiles, and epigenetic modifications [12]. Finally, in clinical development, accurate variant calling enables precise patient stratification for clinical trials based on genetics, leading to smaller, more targeted trials with higher success rates [12].
The connection between variant calling and successful drug development is further strengthened through innovative approaches like patient-derived organoids combined with NGS. This combination allows researchers to study genetic heterogeneity within diseases like cancer and understand how this diversity contributes to drug resistance and poorer outcomes [12]. As drug discovery increasingly embraces personalized medicine, variant calling helps clarify how drugs may affect different patients depending on their genetics, enabling the customization of treatments based on individual genetic profiles [12].
Consequences of Variant Calling Errors in Decision Making
Variant calling errors can significantly derail drug discovery efforts through multiple mechanisms. False positive variants may lead researchers to pursue nonexistent targets, while false negatives can cause genuine therapeutic opportunities to be overlooked [15] [13]. These errors are particularly problematic in complex genomic regions such as homopolymers, segmental duplications, and hard-to-map regions, which often coincide with medically relevant genes [15].
The StratoMod study demonstrated that different variant calling pipelines show substantially different performance characteristics across various genomic contexts [15]. For instance, Illumina excels in low-complexity regions like homopolymers, while Oxford Nanopore Technologies (ONT) shows higher performance in segmental duplications and hard-to-map regions [15]. These contextual performance variations mean that choice of sequencing platform and analysis pipeline can systematically bias which variants are detected, potentially skewing target discovery efforts toward or away from certain genomic regions [15].
In cancer research, where tumor heterogeneity presents additional challenges, variant calling errors can lead to mischaracterization of tumor evolution and drug resistance mechanisms [12] [13]. In pharmacogenomics, errors in identifying genetic variations that affect drug absorption, distribution, metabolism, and excretion can compromise personalized dosing strategies [12]. The cumulative impact of these errors extends beyond scientific missteps to significant financial costs, as misguided clinical trials based on inaccurate genetic information can waste millions of dollars and delay life-saving treatments [12].
Figure 1: Impact of variant calling on drug discovery pipeline. Errors introduce failures (red) while best practices improve outcomes (green).
Comparative Analysis of Variant Calling Approaches
Traditional vs. AI-Enhanced Variant Calling Methods
Variant calling methodologies have evolved significantly from early statistical approaches to modern artificial intelligence (AI)-enhanced methods [16]. Traditional variant callers typically employ rule-based algorithms that apply predetermined thresholds and statistical models to identify genetic variants [16]. While these methods have served as the foundation of genomic analysis for years, they often struggle with complex genomic regions, repetitive sequences, and challenging variant types [16].
AI-enhanced approaches, particularly those utilizing deep learning (DL), represent a paradigm shift in variant calling [16]. These methods leverage convolutional neural networks (CNNs) trained on large-scale genomic datasets to identify subtle patterns that distinguish true variants from sequencing artifacts [16]. Unlike traditional methods that require manual parameter tuning and filtering, DL-based callers can automatically produce filtered variants, eliminating the need for post-calling refinement in many cases [16]. This automation not only improves accuracy but also reduces the bioinformatics expertise required for reliable variant detection [16].
The performance advantage of AI-enhanced methods is particularly evident in challenging genomic contexts. DeepVariant, for example, analyzes pileup image tensors of aligned reads, effectively transforming the variant calling problem into an image recognition task [16]. This approach has demonstrated superior accuracy compared to traditional methods including SAMTools, Strelka, GATK, and FreeBayes [16]. Similarly, Clair3 employs deep learning to achieve better performance, especially at lower coverages traditionally more prone to errors [16]. These advancements are crucial for drug discovery applications where comprehensive and accurate variant detection is essential for target identification.
Benchmarking Performance Across Platforms and Methodologies
Recent comprehensive benchmarking studies provide quantitative evidence of variant caller performance across different sequencing technologies and genomic contexts. A systematic evaluation of state-of-the-art variant calling pipelines tested 45 different combinations of read alignment and variant calling tools on 14 gold standard samples from the Genome in a Bottle (GIAB) consortium [17]. The results revealed "surprisingly large differences in the performance of cutting-edge tools even in high confidence regions of the coding genome," highlighting the critical importance of tool selection [17]. In this extensive benchmark, DeepVariant "consistently showed the best performance and the highest robustness," while other actively developed tools including Clair3, Octopus, and Strelka2 also performed well but with greater dependence on input data quality and type [17].
Table 1: Performance Comparison of Selected Variant Calling Tools
| Variant Caller | Technology Base | SNV Accuracy | Indel Accuracy | Strengths | Limitations |
|---|---|---|---|---|---|
| DeepVariant [16] [17] | Deep Learning (CNN) | >99% [18] | >96% [18] | High accuracy, robust across regions | High computational cost [16] |
| DNAscope [16] [19] | Machine Learning | >99% [18] | >96% [18] | Computational efficiency, no manual filtering | ML-based rather than deep learning [16] |
| GATK [17] | Statistical model | Varies by version | Varies by version | Well-established, extensive documentation | Requires complex filtering [13] |
| Clair3 [17] [20] | Deep Learning | High [20] | High [20] | Fast, excellent at lower coverages | Performance varies by data type [17] |
| Strelka2 [17] | Statistical model | High | High | Good performance | Less robust than DL methods [17] |
The transition from traditional to AI-enhanced variant calling methods shows clear performance benefits. A 2025 study focusing on bacterial genomics demonstrated that deep learning-based variant callers, particularly Clair3 and DeepVariant, "significantly outperform traditional methods and even exceed the accuracy of Illumina sequencing" when applied to Oxford Nanopore Technologies' super-high accuracy model [20]. This superior performance was attributed to the ability of these methods to overcome Illumina's errors, which often arise from difficulties in aligning reads in repetitive and variant-dense genomic regions [20].
For drug discovery applications, the choice between short-read and long-read sequencing technologies also significantly impacts variant calling accuracy. Short-read technologies like Illumina excel in SNV detection and offer cost-effective sequencing, while long-read technologies from PacBio and Oxford Nanopore provide advantages for structural variant detection and resolving complex genomic regions [11] [13]. The emerging approach of hybrid analysis, which combines short-read and long-read data from the same sample, demonstrates promising improvements in variant calling accuracy [19]. The DNAscope Hybrid pipeline, for instance, "significantly improves SNP and Indel calling accuracy, particularly in complex genomic regions," and at lower long-read depths (5x-10x) can outperform standalone short- or long-read pipelines at full sequencing depths (30x-35x) [19].
Table 2: Sequencing Strategy Impact on Variant Calling
| Sequencing Strategy | Target Space | Read Length | SNV/Indel Detection | Structural Variant Detection | Best Applications in Drug Discovery |
|---|---|---|---|---|---|
| Whole Genome Sequencing [14] | ~3200 Mbp | Varies | Excellent | Excellent | Comprehensive variant discovery, novel target identification |
| Whole Exome Sequencing [14] [18] | ~50 Mbp | Short | Excellent | Limited | Coding region focus, cost-effective target screening |
| Targeted Panels [14] | ~0.5 Mbp | Short | Outstanding for low-frequency variants | Limited | Specific gene families, clinical validation |
| Long-Read Sequencing [11] | Full genome | 10,000-30,000 bp | Good | Outstanding | Complex regions, structural variation, repetitive elements |
| Hybrid Approach [19] | Full genome | Combined | Excellent | Excellent | Comprehensive discovery where accuracy is critical |
Experimental Protocols for Optimal Variant Detection
Best Practices in Sample Preparation and Sequencing
The foundation of accurate variant calling begins long before computational analysis, with proper sample preparation and sequencing strategies significantly influencing downstream results [13]. Sample quality deserves particular attention, as degraded or damaged DNA—commonly encountered with formalin-fixed paraffin-embedded (FFPE) samples—can introduce artifacts that complicate variant calling and make it difficult to distinguish between true and damage-induced low-frequency mutations [13]. The use of repair enzymes can help mitigate these issues by removing a broad range of damage, thereby increasing confidence in variant calls [13].
Sequencing strategy selection represents another critical decision point. As shown in Table 2, different approaches offer distinct advantages for various applications in drug discovery [14]. Whole genome sequencing provides the most comprehensive variant detection but at higher cost, while targeted panels offer cost-effective focused analysis with superior sensitivity for low-frequency variants due to higher sequencing depths [14] [13]. The choice between short-read and long-read technologies should align with research objectives, with long-read sequencing particularly valuable for regions inaccessible to short reads [11] [13].
Experimental design should also account for specific variant types of interest. For somatic variant detection in cancer studies, sequencing multiple samples from the same individual increases specificity, helping distinguish true somatic variants from artifacts [13]. Similarly, for familial disorders, trio sequencing (child and both parents) enhances accuracy by providing genetic context [16]. Each of these considerations must be balanced against practical constraints including cost, sample availability, and downstream analysis capabilities.
Bioinformatics Pipelines and Quality Control
Implementing robust bioinformatics pipelines is essential for accurate variant calling, with several critical steps required before variant detection itself [14]. The process typically begins with read alignment using tools such as BWA-MEM, Bowtie2, or minimap2, which map raw sequencing reads to a reference genome [14]. During this stage, prioritizing sensitivity over specificity ensures potential variants are not overlooked initially [13]. Following alignment, identifying and marking PCR duplicates—redundant reads originating from the same nucleic acid molecule—helps prevent overcounting of amplification artifacts [14]. Tools like Picard Tools or Sambamba are commonly used for this purpose [14].
Quality control represents a crucial but sometimes overlooked component of the variant calling pipeline [14]. Routine QC of analysis-ready BAM files should evaluate key sequencing metrics, verify sufficient sequencing coverage was achieved, and check for sample contamination [14]. For family studies and paired samples, expected relationships should be confirmed using tools like the KING algorithm [14]. Additional processing steps such as base quality score recalibration (BQSR) and local realignment around indels may be implemented, though evaluations suggest these provide marginal improvements for their computational cost [14].
The selection of appropriate benchmarking resources enables objective evaluation of variant calling performance [14] [18]. The Genome in a Bottle (GIAB) consortium provides gold standard datasets with high-confidence variant calls for several well-characterized genomes, allowing researchers to compare their results against established benchmarks [14] [18]. These resources facilitate the calculation of standard performance metrics including precision, recall, and F1 scores, providing quantitative measures of variant calling accuracy [18]. For clinical applications, the Association for Molecular Pathology recommends using representative variants in bioinformatics guidelines, which must satisfy regulatory requirements for submissions [15].
Figure 2: Optimal variant calling workflow. Critical pre-calling considerations in yellow.
Table 3: Essential Research Reagent Solutions for Variant Calling
| Resource | Function | Application in Drug Discovery |
|---|---|---|
| GIAB Reference Materials [14] [18] | Gold standard genomes for benchmarking | Validating variant calling pipeline performance |
| Agilent SureSelect Kits [18] | Exome capture and library preparation | Target enrichment for coding region focus |
| FFPE DNA Repair Mix [13] | DNA damage reversal in archived samples | Enabling variant calling from clinical specimens |
| Patient-Derived Organoids [12] | Disease modeling using human cells | Studying genetic heterogeneity and drug response |
| Corning Organoid Culture Products [12] | Specialized surfaces and media | Maintaining genetically stable disease models |
The critical importance of variant calling accuracy in drug target discovery cannot be overstated. As this comparison guide has demonstrated, errors in variant detection can fundamentally misdirect research efforts, leading to missed therapeutic opportunities and costly failed clinical trials. The emergence of AI-enhanced variant calling methods represents a significant advancement over traditional approaches, with tools like DeepVariant, DNAscope, and Clair3 consistently demonstrating superior performance in benchmarking studies [18] [16] [17].
Successful implementation of variant calling in drug discovery requires attention to the entire workflow—from sample preparation through computational analysis. The choice of sequencing strategy should align with research objectives, with hybrid approaches offering particular promise for comprehensive variant detection [19]. As the field advances, the development of more diverse gold standard genomes and improved benchmarking in challenging genomic regions will further enhance variant calling accuracy [17].
For researchers and drug development professionals, investing in optimal variant calling practices is not merely a technical consideration but a fundamental requirement for success. By adopting the best practices, tools, and validation frameworks outlined in this guide, the drug discovery community can significantly improve the reliability of target identification and validation, ultimately accelerating the development of more effective, personalized therapies.
In chemogenomic screens, where researchers systematically study the interactions between chemical compounds and biological systems, the accurate detection of genetic variants through Next-Generation Sequencing (NGS) is paramount. These screens rely on identifying true compound-induced genetic changes amidst technical noise to understand drug mechanisms, identify resistance markers, and discover new therapeutic targets. The reliability of these findings, however, is continually challenged by three persistent technical pitfalls: PCR-derived artifacts, alignment ambiguities, and the inherent difficulties in detecting low-frequency variants. These challenges are particularly pronounced in clinical sequencing contexts, where false positives can lead to incorrect therapeutic decisions, and false negatives can miss biologically significant mutations present in subpopulations of cells [14]. This guide objectively compares the performance of various experimental and computational approaches for mitigating these challenges, providing researchers with data-driven insights to optimize their variant calling accuracy in chemogenomic research.
PCR Artifacts: Origins and Mitigation Strategies
PCR artifacts introduced during library preparation represent a major source of false-positive variant calls. These errors are not merely stochastic but can arise from specific, identifiable mechanisms. One significant source is the oxidation of DNA during fragmentation, particularly during acoustic shearing. This process can generate 8-oxoguanine (8-oxoG) lesions, which subsequently cause C>A/G>T transversion artifacts during sequencing. These artifacts are characterized by their presence at low allelic fractions, specific strand orientation (G>T errors in the first Illumina read, C>A in the second), and occurrence in both tumor and normal samples, indicating a non-biological origin [21]. Another source involves the generation of chimeric reads during library fragmentation. Studies comparing ultrasonic and enzymatic fragmentation have revealed that sonication can create artifacts containing inverted repeat sequences (IVSs), while enzymatic fragmentation tends to produce artifacts centered on palindromic sequences (PS) with mismatched bases. These chimeric molecules are formed through a mechanism termed the PDSM model (pairing of partial single strands derived from a similar molecule), where sheared DNA fragments incorrectly reanneal [22].
Experimental Protocols for Artifact Reduction
Antioxidant Supplementation Protocol: To mitigate oxidation artifacts during DNA shearing, researchers can introduce antioxidant agents to the DNA sample before acoustic shearing. The following protocol, adapted from Costello et al., has proven effective [21]:
- Perform a solid-phase reversible immobilization (SPRI) bead cleanup (e.g., using Ampure XP beads) on genomic DNA to remove contaminants from extraction.
- Elute the DNA in 50 µL of an antioxidant-supplemented buffer. Tested conditions include:
- Condition A: 10 mM Tris-HCl + 1 mM EDTA
- Condition B: 10 mM Tris-HCl + 100 µM Deferoxamine Mesylate (DFAM)
- Condition C: 10 mM Tris-HCl + 100 µM Butylated Hydroxytoluene (BHT)
- Condition D: A combination of all three antioxidants.
- Proceed with standard Covaris shearing and subsequent library preparation steps. ELISA-based quantification of 8-oxoG levels can confirm reduction of oxidative damage.
Unique Molecular Identifier (UMI) Integration Workflow: UMIs are short random nucleotide sequences ligated to DNA fragments before any PCR amplification steps. This allows bioinformatic consensus generation to distinguish true original molecules from PCR errors [23]. A typical UMI workflow using the fgbio toolkit involves:
- Annotate Bam with Umis: Tag reads in a BAM file with their UMI sequences.
- Group Reads by UMI: Cluster reads that share both a UMI and mapping coordinates into "UMI families."
- Create Consensus Reads: Generate a single, high-quality consensus sequence for each UMI family. Variants not present in the consensus of a family are considered PCR errors and filtered out. Table 1: Comparison of Antioxidant Efficacy in Reducing Oxidation Artifacts (Based on Costello et al. [21])
| Antioxidant Condition | Relative Reduction in C>A Artifacts | Key Observation |
|---|---|---|
| 1 mM EDTA | Moderate | Chelates metal ions that catalyze oxidation. |
| 100 µM DFAM | High | Potent iron chelator, highly effective. |
| 100 µM BHT | Moderate | Lipid-soluble antioxidant. |
| Combination (All three) | Highest | Synergistic effect, most comprehensive protection. |
Performance Comparison of UMI-Aware Variant Callers
The use of UMIs necessitates specialized variant callers. A 2024 benchmark study compared six variant callers on ctDNA data, including two UMI-aware callers [23]. Table 2: Benchmarking of Variant Callers on Low-Frequency Variants (Synthetic Data) [23]
| Variant Caller | Type | Reported Sensitivity | Reported Specificity | Key Finding |
|---|---|---|---|---|
| UMI-VarCal | UMI-aware | High | Highest | Detected the fewest putative false positives in UMI data. |
| Mutect2 | Standard | Highest | Medium (Low without UMIs) | Balanced sensitivity/specificity with UMIs; high false positives without. |
| LoFreq | Standard | High | Medium | Effective for low-frequency calls but susceptible to PCR artifacts. |
| bcftools | Standard | Medium | High | Conservative caller, may miss true low-frequency variants. |
| FreeBayes | Standard | Medium | Medium | Balanced performance but outperformed by UMI-aware methods. |
The data indicates that while standard callers like Mutect2 can achieve high sensitivity, they tend to generate more privately called variants—a potential indicator of false positives—in data without UMIs. The integration of UMIs with UMI-aware callers like UMI-VarCal provides a superior balance for distinguishing true low-frequency variants from PCR artifacts [23].
Diagram 1: UMI Workflow for PCR Error Correction. This diagram illustrates the process of using Unique Molecular Identifiers (UMIs) to tag original DNA molecules before PCR. Bioinformatic grouping into UMI families allows the generation of a consensus sequence, which effectively filters out stochastic PCR errors that are not present in the majority of reads within a family.
Alignment Ambiguity: Impact on Variant Calling Fidelity
Challenges in Spliced Alignment for RNA Sequencing
Variant calling from RNA-seq data presents unique alignment challenges not encountered in DNA-seq. The primary issue stems from the presence of introns in pre-mRNA, which results in sequencing reads that are "spliced" when aligned to a reference genome. These spliced alignments contain large gaps (represented by 'N' in the CIGAR string of the BAM file), which disrupt the contiguous read pileups that DNA-based variant callers are designed to analyze [24]. This misalignment between data structure and tool expectation leads to two major problems: reduced sensitivity (false negatives) and compromised precision (false positives), particularly for variants near exon-intron boundaries.
Optimized Protocol for lrRNA-Seq Variant Calling
A 2023 study demonstrated that transforming alignment files is critical for achieving high performance with DNA-based variant callers on long-read RNA sequencing (lrRNA-seq) data. The recommended pipeline for tools like DeepVariant is as follows [24]:
- SplitNCigarReads (SNCR): Use the GATK function to split reads at intron gaps (N in CIGAR). This converts a single long read spanning multiple exons into several shorter, contiguous alignment segments.
- flagCorrection: A critical, often overlooked step. The SNCR tool assigns the "primary alignment" flag to only one segment from a split read, marking others as "supplementary." The custom
flagCorrectiontool resets all fragments from the original read to be primary alignments, preventing their accidental filtration by downstream tools. - Variant Calling with DeepVariant: Process the transformed BAM file with DeepVariant, which uses a deep learning model to call variants from the now-contiguous read pileups.
Performance benchmarks on PacBio Iso-Seq data from Jurkat and WTC-11 cell lines showed that this combined SNCR + flagCorrection + DeepVariant pipeline significantly outperformed using DeepVariant on unmodified BAMs or with SNCR alone, especially in regions with low-to-moderate read coverage (≤ 40x) and a high proportion of intron-containing reads [24].
The Low-Allele-Frequency Challenge: Distinguishing Signal from Noise
The Fundamental Detection Limit Problem
The reliable detection of mutations present at very low frequencies is crucial for chemogenomics, where it can reveal rare resistant subclones or the early effects of a compound. The core challenge is that the expected frequency of true biological variants (e.g., ~10⁻⁸ to 10⁻⁵ mutations per nucleotide for independent events) falls far below the background error rate of standard Illumina sequencing (~5 × 10⁻³ per nucleotide) [25]. Without specialized methods, even variants with a Variant Allele Frequency (VAF) of 0.5% - 1% are often spurious. Factors that can push a true variant above this noise floor include DNA damage hyperhotspots, clonal expansion of a mutant cell, or analyzing very small biopsies [25].
Advanced Methods for Ultra-Sensitive Detection
To breach this barrier, several sophisticated sequencing methods have been developed, primarily relying on consensus strategies. These can be categorized based on how they use the original template strand information [25]:
- Single-Strand Consensus Sequencing (SSCS): Methods like Safe-SeqS and SiMSen-Seq use UMIs to group reads derived from the same original single DNA strand. Errors are reduced by requiring a consensus within these groups.
- Duplex Sequencing (DS): Ultrasensitive methods like DuplexSeq, SaferSeq, and NanoSeq tag and sequence both strands of the original DNA duplex independently. A true variant is only called if it is found in the consensus sequences derived from both complementary strands. This approach can push the error rate down to ~10⁻⁹ per nucleotide, as it corrects for errors arising from DNA damage on a single strand [25].
SPIDER-seq Protocol for PCR-based Libraries: A novel method called SPIDER-seq (2025) addresses the challenge of applying UMIs to general PCR-based libraries, where UMI sequences are overwritten in subsequent cycles. Its protocol is [26]:
- Amplification with UID-Primers: Perform multiple cycles (e.g., 6 cycles) of PCR using primers containing random Unique Identifiers (UIDs).
- Peer-to-Peer Network Clustering: Bioinformatically cluster all daughter molecules derived from a single original strand by constructing a network based on shared UIDs between parental and daughter strands. This creates a Cluster Identifier (CID).
- CID-Based Consensus Generation: Generate a high-fidelity consensus sequence for each CID, effectively correcting for sequencing and late-cycle PCR errors.
SPIDER-seq has demonstrated the ability to detect mutations at frequencies as low as 0.125% from amplicon libraries, offering a more cost-effective and rapid alternative to hybridization-capture-based UMI methods for applications like monitoring a defined set of mutations [26].
Informatics-Based Filtering for Low-VAF Variants
In the absence of wet-lab consensus methods, robust bioinformatic filtering is essential. Key strategies include:
- Variant Allele Frequency (VAF) Cutoffs: Empirical data from clinical exome sequencing suggests that setting a VAF cutoff at approximately 0.30 (30%) can filter out a significant portion (∼82%) of technical artifacts while retaining all medically relevant heterozygous variants, which are expected at VAFs between 0.33 and 0.63 [27]. For somatic cancer variants or mosaic germline variants, lower, validated cutoffs must be established.
- Artifact Blacklisting: Tools like
ArtifactsFindercan systematically scan a BED target region to identify locations prone to artifacts from inverted repeats (IVSs) or palindromic sequences (PSs), generating a custom "blacklist" to filter false positives [22]. - Orientation Filtering: For oxidation artifacts, a strong indicator is a strand bias where G>T errors are found exclusively in read 1 and C>A errors in read 2. Filtering variants that display this pattern can effectively remove these artifacts [21].
Table 3: Comparison of Ultrasensitive Variant Detection Methods [25]
| Method Category | Example Methods | Key Principle | Reported Sensitivity | Advantages | Limitations |
|---|---|---|---|---|---|
| Single-Strand Consensus | Safe-SeqS, SiMSen-Seq | Consensus from multiple reads of one original strand. | VAF ~10⁻⁵ | Good error reduction; simpler than duplex. | Cannot correct for single-strand DNA damage. |
| Duplex Sequencing | DuplexSeq, SaferSeq | Independent consensus from both strands of DNA duplex. | MF <10⁻⁹ per nt | Highest accuracy; corrects for strand damage. | More complex; lower library yield. |
| Amplicon with UID Overwriting | SPIDER-seq | Network clustering of PCR reads with overwritten UIDs. | VAF ~0.125% | Cost-effective; fast; suitable for amplicons. | New method; requires specialized pipeline. |
The Scientist's Toolkit: Essential Reagents and Computational Tools
Table 4: Research Reagent Solutions for NGS Variant Calling
| Reagent / Tool | Function / Purpose | Key Application |
|---|---|---|
| Antioxidants (EDTA, DFAM, BHT) | Mitrate oxidative DNA damage during acoustic shearing. | Reduction of C>A/G>T transversion artifacts [21]. |
| Unique Molecular Identifiers (UMIs) | Molecular barcoding of original DNA molecules before amplification. | Tagging and tracking molecules to generate consensus sequences and remove PCR errors [23]. |
| Enzymatic Fragmentation Mix | Alternative to sonication for DNA shearing; minimal DNA loss. | Library prep from low-input samples; requires awareness of palindromic sequence artifacts [22]. |
| ArtifactsFinder | Bioinformatic algorithm to identify artifact-prone genomic sites. | Generation of a custom "blacklist" for filtering false positives from fragmentation artifacts [22]. |
| SPIDER-seq Pipeline | Computational tool for clustering reads and generating CIDs. | Enables ultra-sensitive variant calling from standard PCR amplicons [26]. |
| FlagCorrection Tool | Corrects alignment flags after splitting spliced RNA-seq reads. | Critical pre-processing step for accurate variant calling from lrRNA-seq data using DNA-based callers [24]. |
Diagram 2: Integrated Strategy for Overcoming NGS Pitfalls. This diagram outlines the multi-faceted approach required for accurate variant calling, combining wet-lab experimental techniques, alignment file pre-processing, and rigorous bioinformatic filtering to address the intertwined challenges of PCR artifacts, alignment ambiguity, and low-frequency variants.
Navigating the pitfalls of PCR artifacts, alignment ambiguity, and low-frequency variants requires an integrated strategy combining rigorous wet-lab protocols with sophisticated bioinformatic tools. The experimental data and comparisons presented in this guide demonstrate that no single tool or method is universally superior; rather, the choice depends on the specific application, sample type, and available resources. For instance, while Duplex Sequencing offers the highest theoretical accuracy, SPIDER-seq provides a powerful and more accessible alternative for amplicon-based screens. Critically, the baseline performance of any variant calling pipeline must be established using validated reference standards with known, spiked-in variants to quantify sensitivity and specificity accurately [28]. For chemogenomic screens, where the accurate identification of genetic variants directly impacts the interpretation of compound mechanism and efficacy, adopting these best practices is not merely an optimization but a necessity for generating reliable and actionable data.
Building Robust Pipelines: Best Practices and Tool Selection for Chemogenomic Data
In the field of chemogenomics, where understanding the genetic basis of drug response is paramount, the accuracy of next-generation sequencing (NGS) variant calling is a critical foundation for reliable research outcomes. The choice of computational tools in the bioinformatics pipeline—specifically the aligner and variant caller—directly impacts the sensitivity and precision of variant discovery, which in turn influences downstream analyses and conclusions. This guide provides an objective, data-driven comparison of two widely used aligners, BWA-MEM and Bowtie2, and three established variant callers—GATK, Samtools (Bcftools), and Freebayes—to help researchers and drug development professionals select the optimal tools for their projects.
Aligner Performance: BWA-MEM vs. Bowtie2
BWA-MEM (Burrows-Wheeler Aligner - Maximal Exact Matches) is designed for aligning sequencing reads of 100 bp and longer. It uses a seed-and-extend approach, employing an affine-gap Smith-Waterman algorithm for extension and implementing heuristics to avoid extending alignments through poorly mapping regions. It is particularly well-suited for handling a wide range of read lengths (up to 1 Mbp) and performs chimeric alignments [29] [30].
Bowtie2 is an ultrafast, memory-efficient tool for aligning sequencing reads. It uses the FM-index for efficient sequence search and typically operates in one of two modes: a fast, end-to-end mode (BT-E2E) ideal for reads expected to align entirely to the reference, or a more sensitive local alignment mode (BT-LOC) that allows for partial alignments of reads, which can be beneficial for reads with adapter sequence or significant polymorphisms [17].
Performance Comparison Data
The table below summarizes key performance characteristics as established in benchmarking studies.
Table 1: Performance Comparison of BWA-MEM and Bowtie2
| Feature | BWA-MEM | Bowtie2 | Experimental Context |
|---|---|---|---|
| General Accuracy | Consistently high; often a gold standard in benchmarks [17] | Lower F1 scores in some benchmarks; BT-LOC mode can be more sensitive than BT-E2E [17] | Evaluation using GIAB gold standard samples for variant discovery in coding sequences [17] |
| Alignment Speed | Faster (e.g., ~1042 sec for 2GB FASTQ) [31] | Slower (e.g., ~5132 sec for same dataset) [31] | Empirical test with paired-end FASTQ files [31] |
| Alignment Specificity | High; can be optimized for multi-species samples by increasing seed length [30] | Information not available in search results | Analysis of host-pathogen (e.g., Plasmodium-human) data; default seed length is 19nt [30] |
| Recommended Use Case | General-purpose alignment for WGS and WES; preferred for medical variant calling [17] | Not recommended for medical variant calling in one benchmark; may be suitable for other sequencing applications [17] | Benchmark of state-of-the-art variant calling pipelines [17] |
Variant Caller Performance: GATK, Samtools/Bcftools, and Freebayes
GATK (Genome Analysis Toolkit) HaplotypeCaller is a widely adopted, complex tool that operates by reassembling reads in regions of potential variation. It uses a pair-hidden Markov model for local reassembly of haplotypes and a powerful Markov model-based genotyping algorithm to calculate genotype likelihoods. Its Best Practices pipeline includes additional steps like base quality score recalibration (BQSR) and variant quality score recalibration (VQSR) to refine results [32] [17].
Samtools/Bcftools is a pipeline that relies on the mpileup command to summarize read alignments and compute genotype likelihoods at each genomic position. The bcftools call command then performs the actual variant calling. It is known for its speed and efficiency and can be run in both single-sample and multiple-sample (joint-calling) modes [32].
Freebayes is a Bayesian genetic variant caller that detects polymorphisms—SNPs, indels, mnps, and complex events—by counting the observed alleles and assigning a probability based on their frequency in the population of aligned reads. It is a straightforward method that assumes diploidy but does not rely on complex machine learning models [32].
Performance Comparison Data
The table below summarizes the performance of these callers based on recent benchmarking studies.
Table 2: Performance Comparison of GATK, Bcftools, and Freebayes
| Variant Caller | SNP F1 Score (Example) | Indel F1 Score (Example) | Key Strengths & Weaknesses |
|---|---|---|---|
| GATK HaplotypeCaller | High (e.g., >99% concordance with truth sets) [33] | High | Strengths: Highly polished, extensive best practices, good overall accuracy. Weaknesses: Can be computationally slow, complex to set up [33]. |
| Bcftools | High sensitivity, especially in multiple-sample mode [32] | Information not available in search results | Strengths: Very fast, high specificity, efficient for large projects. Weaknesses: May have lower sensitivity in single-sample mode at low coverage [32]. |
| Freebayes | Lower number of detected variants in some comparisons [32] | Information not available in search results | Strengths: Simple, model-free approach. Weaknesses: May have lower sensitivity and specificity compared to other methods [32]. |
| DeepVariant | Highest in benchmarks (e.g., >99.9%) [17] | High (e.g., >99.5%) [3] | Strengths: Top-tier accuracy and robustness using deep learning. Weaknesses: Computationally intensive [17]. |
Integrated Workflows and Experimental Protocols
Standardized Benchmarking Methodology
To ensure fair and reproducible comparisons, benchmarking studies often follow a rigorous protocol based on gold-standard reference materials.
- Reference Datasets: The Genome in a Bottle (GIAB) consortium provides well-characterized human genomes (e.g., NA12878/HG001, Ashkenazi Jewish trio HG002-HG004) with high-confidence variant calls that serve as the "truth set" for evaluation [34] [17].
- Data Processing:
- Alignment: Raw FASTQ reads are aligned to a reference genome (GRCh37/hg19 or GRCh38) using the aligner with default or optimized parameters [17].
- Post-Alignment Processing: The resulting BAM files are processed to mark duplicate reads (e.g., using GATK MarkDuplicates) and, for some pipelines, to perform Base Quality Score Recalibration (BQSR) [35] [17].
- Variant Calling: Processed BAM files are used as input for the variant callers.
- Performance Assessment: The generated VCF files are compared against the GIAB truth set using the
hap.pytool. Key metrics include:
Impact of Alignment on Variant Calling
A critical finding from recent research is that while the choice of aligner is important, its impact is often superseded by the choice of the variant caller, provided a robust aligner like BWA-MEM is used. One large-scale study concluded that when considering accurate aligners (excluding Bowtie2, which performed poorly), "the accuracy of variant discovery mostly depended on the variant caller and not the read aligner" [17]. However, the alignment and variant calling steps are not entirely independent. The DRAGEN platform, which uses a highly optimized alignment algorithm, demonstrated systematically higher F1 scores, precision, and recall compared to a GATK with BWA-MEM2 pipeline, underscoring that improvements in the alignment stage can translate to better final variant calls [34].
The following diagram illustrates the standard workflow for benchmarking aligners and variant callers.
Essential Research Reagent Solutions
This table lists key computational tools and resources that form the backbone of a reliable NGS variant calling workflow.
Table 3: Key Research Reagents and Resources for NGS Variant Calling
| Resource Name | Type | Primary Function in Workflow |
|---|---|---|
| GIAB Reference Samples | Benchmark Dataset | Provides a gold-standard set of genomes with expertly curated variant calls to validate pipeline accuracy [34]. |
| GRCh37/hg38 | Reference Genome | The standard human reference sequences to which reads are aligned for mapping and variant identification. |
| BWA-MEM | Read Aligner | Aligns sequencing reads to the reference genome, a critical first step that influences all downstream analysis [29] [17]. |
| GATK | Variant Calling Toolkit | A comprehensive suite of tools, with HaplotypeCaller being a benchmark for accurate germline variant discovery [17]. |
| DeepVariant | Variant Caller | A deep-learning based caller that has demonstrated top-tier accuracy in independent benchmarks [3] [17]. |
| SAMtools/Bcftools | Utility Suite | A collection of utilities for manipulating alignments and calling variants, prized for its speed and efficiency [32]. |
| hap.py | Benchmarking Tool | The official GA4GH tool for calculating performance metrics like precision and recall against a truth set [17]. |
The choice between BWA-MEM and Bowtie2 is clear-cut for variant calling applications: BWA-MEM is the recommended aligner due to its superior performance in benchmarking studies, higher speed, and status as a de facto gold standard in medical genomics [17]. For variant calling, the landscape is more nuanced. While GATK HaplotypeCaller remains a robust and widely supported option with very high accuracy, newer tools like DeepVariant have demonstrated superior performance in recent, comprehensive benchmarks [17].
For researchers in chemogenomics, where accuracy is non-negotiable, the evidence suggests a pipeline combining BWA-MEM for alignment and DeepVariant for calling would yield the most accurate results. If computational resources or support for DeepVariant are a constraint, the established BWA-MEM and GATK HaplotypeCaller pipeline remains a very strong alternative. Tools like Bcftools are excellent for projects where processing speed is a critical factor, and it has been shown to outperform other callers in specific scenarios, such as low-coverage data or multiple-sample calling modes [32].
In chemogenomic screens, the precise identification of variants is not merely a preliminary step but the foundation upon which all subsequent analyses and therapeutic insights are built. The central challenge in this process lies in the accurate discrimination between somatic mutations, which are acquired and specific to the tumor cells, and germline variants, which are inherited and present in all of a patient's cells. This distinction is critical because somatic mutations can drive cancer progression and dictate response to therapies, whereas germline variants provide the constitutional genetic background of the individual. The failure to properly separate these variant types directly compromises the integrity of chemogenomic data, leading to misinterpretation of drug response biomarkers and potentially flawed therapeutic associations.
Next-generation sequencing (NGS) technologies have become the standard for variant detection in cancer research, yet each platform presents distinct advantages and limitations for chemogenomic applications. Short-read sequencing (e.g., Illumina) currently offers higher base-level accuracy and is widely adopted in clinical settings, but struggles with highly homologous genomic regions, including paralogous genes and pseudogenes, which can lead to false positives or negatives [36]. Conversely, emerging long-read sequencing technologies (PacBio, Nanopore) excel in resolving complex genomic regions and providing phasing information, making them particularly valuable for pharmacogenes with structural complexity, such as CYP2D6, CYP2B6, and HLA genes [37]. The choice of sequencing technology must align with the specific genomic contexts of the drug targets under investigation in chemogenomic screens.
Fundamental Biological and Technical Distinctions
Biological Origins and Clinical Implications
Somatic and germline variants originate through fundamentally different biological mechanisms and have distinct implications for cancer biology and treatment.
Somatic Variants: These mutations occur in non-germline tissues after conception and are not inherited. In cancer, somatic mutations accumulate due to environmental exposures, replication errors, and defective DNA repair mechanisms. They are present only in tumor cells and their progeny, leading to mosaicism within tissues. From a clinical perspective, somatic mutations in genes such as BRAF, EGFR, and KRAS can serve as direct therapeutic targets or predictive biomarkers for drug response. Identifying these variants helps guide targeted therapies and is essential for calculating clinically relevant metrics like tumor mutational burden (TMB), an important predictor of response to immunotherapy [38].
Germline Variants: These are inherited genetic variations present in virtually every cell from birth. While most are benign polymorphisms, pathogenic germline variants in cancer predisposition genes (e.g., BRCA1, BRCA2, TP53) confer increased lifetime risk of developing specific malignancies. In chemogenomic contexts, germline variants in pharmacogenes can significantly influence drug metabolism, efficacy, and toxicity risk. For instance, polymorphisms in genes like CYP2C9, CYP2C19, and DPYD affect the metabolism of numerous chemotherapeutic agents and targeted therapies [37].
The accurate classification of these variant types is not merely an academic exercise but has direct clinical consequences. Misclassification can lead to inappropriate treatment decisions, miscalculated TMB scores, and incorrect assessment of hereditary cancer risk. This is particularly challenging in tumor-only sequencing designs, where the absence of a matched normal sample complicates the discrimination between somatic and germline variants [38].
Technical Challenges in Variant Discrimination
Several technical factors complicate the accurate distinction between somatic and germline variants in sequencing data:
Germline Leakage: This occurs when germline variants are mistakenly identified as somatic mutations due to limitations in variant calling algorithms. The median somatic SNV prediction set contains approximately 4325 calls and leaks about one germline polymorphism, with leakage rates inversely correlated with somatic SNV prediction accuracy [39]. This leakage poses privacy concerns as leaked germline variants could potentially be used for patient re-identification.
Tumor-in-Normal Contamination: The unexpected contamination of normal samples with tumor cells reduces variant detection sensitivity, compromising downstream analyses. This problem is particularly prevalent in haematological malignancies and sarcomas, with highest prevalence observed in saliva samples from acute myeloid leukaemia patients and sorted CD3+ T-cells from myeloproliferative neoplasms [40]. Such contamination can lead to erroneous subtraction of genuine high-allele-frequency somatic variants during variant calling.
Mapping Artifacts in Homologous Regions: Short-read sequencing technologies face significant challenges in highly homologous genomic regions such as pseudogenes or paralogous genes. Genes with high homology (e.g., SMN1, SMN2, CBS, and CORO1A) show consistently low coverage across all read lengths due to nonspecific mapping, potentially leading to false negative results in critical pharmacogenes [36].
Comparative Performance of Variant Calling Approaches
Individual Caller Performance Benchmarking
Comprehensive benchmarking studies provide critical insights into the relative performance of somatic variant callers, enabling informed selection for chemogenomic applications. A recent evaluation of 20 somatic variant callers across multiple whole-exome sequencing datasets revealed significant differences in accuracy for detecting single-nucleotide variants (SNVs) and insertions/deletions (indels) [41].
Table 1: Performance of Leading Somatic Variant Callers
| Variant Caller | SNV F1 Score | Indel F1 Score | Notable Strengths |
|---|---|---|---|
| Dragen | 0.895 (Highest for SNVs) | - | Commercial solution with optimized performance |
| Mutect2 | ~0.89 | ~0.837 | Widely adopted, balanced SNV/indel performance |
| Muse | ~0.88 | - | High SNV accuracy |
| NeuSomatic | - | 0.837 (Highest for indels) | Deep learning approach |
| TNScope | ~0.87 | - | Commercial solution |
| Strelka | ~0.86 | ~0.82 | Robust open-source option |
| VarScan2 | ~0.80 | ~0.81 | Established method |
The benchmarking study identified five high-performing individual somatic variant callers: Muse, Mutect2, Dragen, TNScope, and NeuSomatic [41]. Performance varied significantly across different reference datasets, highlighting the importance of evaluating callers on datasets representative of specific research contexts. For chemogenomic applications focused on specific pharmacogenes, additional validation in genomic regions relevant to drug metabolism and response is recommended.
Ensemble Approaches for Enhanced Accuracy
Ensemble methods that combine multiple variant callers have demonstrated superior performance compared to individual callers, achieving significantly higher F1 scores for both SNVs and indels [41].
Table 2: High-Performing Ensemble Combinations
| Variant Type | Ensemble Composition | Performance (F1 Score) | Improvement Over Best Single Caller |
|---|---|---|---|
| SNVs | LoFreq, Muse, Mutect2, SomaticSniper, Strelka, Lancet | 0.927 | >3.6% improvement over Dragen |
| Indels | Mutect2, Strelka, Varscan2, Pindel | 0.867 | >3.5% improvement over NeuSomatic |
| Optimal Balanced | Muse, Mutect2, Strelka (SNVs); Mutect2, Strelka, Varscan2 (Indels) | >0.89 (SNVs), >0.85 (Indels) | Cost-effective solution with high accuracy |
The ensemble approach that combined six callers (LoFreq, Muse, Mutect2, SomaticSniper, Strelka, and Lancet) for SNVs achieved a mean F1 score of 0.927, outperforming the top-performing individual caller (Dragen) by more than 3.6% [41]. Similarly, for indels, a four-caller ensemble (Mutect2, Strelka, Varscan2, and Pindel) achieved a mean F1 score of 0.867, representing a 3.5% improvement over the best individual indel caller (NeuSomatic) [41]. These ensemble methods effectively leverage the complementary strengths of individual callers, mitigating their respective limitations.
Tumor-Only Calling and Machine Learning Advances
In clinical settings where matched normal samples are unavailable, tumor-only variant calling presents significant challenges, primarily due to difficulty distinguishing rare germline variants from true somatic mutations. Traditional approaches that rely on filtering against germline databases (e.g., dbSNP, gnomAD) exhibit substantial false positive rates, particularly for patients from populations underrepresented in these databases [38].
Recent machine learning approaches have demonstrated remarkable improvements in tumor-only variant calling. Studies applying TabNet, XGBoost, and LightGBM to classify variants as somatic or germline using features derived exclusively from tumor-only data achieved area under the curve (AUC) values exceeding 94% on TCGA datasets and 85% on metastatic melanoma datasets [38]. These models utilized 30 mutation- and copy-number-specific features, including:
- Traditional features: germline database frequency, COSMIC somatic mutation database counts, variant allele fraction (VAF)
- Sequence context features: trinucleotide context and base substitution subtypes
- Local copy number features: derived from copy-number segmentation data
Notably, these machine learning approaches successfully eliminated the significant racial bias observed in traditional tumor-only variant calling methods, where TMB estimates for Black patients were extremely inflated relative to those of white patients due to underrepresented germline variants in reference databases [38].
Experimental Protocols for Robust Variant Detection
Standardized Sequencing and Analysis Workflow
Implementing a robust, reproducible variant calling pipeline requires strict adherence to standardized protocols across sample processing, sequencing, and bioinformatic analysis. The following workflow represents best practices derived from comprehensive benchmarking studies:
Sample Preparation and Sequencing:
- Utilize matched tumor-normal pairs when possible, with normal tissue derived from blood, saliva, or skin biopsy
- Implement rigorous quality control measures using tools like omnomicsQ to flag samples falling below predefined thresholds [42]
- For WES, ensure minimum coverage of 100× for tumor samples and 75× for normal samples to confidently detect variants with VAFs ≥0.15 [43]
- Consider longer read lengths (150-250 bp) to improve mapping accuracy in homologous regions [36]
Bioinformatic Processing:
- Alignment with BWA-MEM against appropriate reference genome (hg19/hg38)
- Post-alignment processing including duplicate marking (sambamba) and base quality score recalibration (GATK BQSR) [41]
- Multi-caller approach using at least 2-3 high-performing callers (e.g., Mutect2, Strelka, VarScan2)
- Ensemble aggregation of calls with threshold-based filtering
- Annotation using tools like ANNOVAR, Ensembl VEP, or SnpEff [42]
Quality Assurance and Validation:
- Assess tumor-in-normal contamination using tools like TINC [40]
- Apply systematic quality segmentation to identify genomic regions with high false-positive rates [43]
- Validate low-frequency variants in difficult genomic regions with orthogonal methods
Contamination Assessment and Quality Control
Accurate variant calling requires meticulous quality control throughout the analytical process, with particular attention to sample contamination:
Tumor-in-Normal Contamination Assessment:
- Implement the TINC (Tumor IN Normal contamination) method to quantify the percentage of tumor cells in normal samples [40]
- TINC utilizes variant allele frequencies (VAFs) of clonal somatic SNVs detected in both tumor and normal samples
- The method identifies high-confidence clonal mutations using MOBSTER for subclonal deconvolution
- Expected performance: R² = 0.95 for haematological cancers and R² = 0.85 for lung cancers in synthetic validation [40]
Systematic Quality Segmentation:
- Leverage cohort-level metrics to identify genomic regions with systematically high or low quality [43]
- Aggregate base quality, mapping quality, and depth metrics across multiple samples
- Label approximately 90% of non-N autosomal regions as high-quality and 10% as low-quality
- This approach captures 86-89% of false-positive SNVs in low-quality regions, enabling targeted filtering [43]
Essential Research Reagents and Computational Tools
The Scientist's Toolkit for Variant Detection
Table 3: Essential Research Reagents and Computational Tools
| Category | Tool/Reagent | Primary Function | Considerations for Chemogenomics |
|---|---|---|---|
| Variant Callers | Mutect2 | Somatic variant detection | High balanced performance for SNVs/indels; part of GATK |
| Strelka | Somatic variant detection | Fast, accurate; good for low-frequency variants | |
| VarScan2 | Somatic variant detection | Established method; good for heterogeneous tumors | |
| Dragen | Integrated pipeline | Commercial; hardware-accelerated; high accuracy | |
| Ensemble Methods | Custom combinations | Aggregate multiple callers | Optimal: Muse+Mutect2+Strelka (SNVs); Mutect2+Strelka+Varscan2 (indels) [41] |
| Quality Control | TINC | Tumor-in-normal contamination | Essential for leukaemia, sarcoma; uses VAF distributions [40] |
| GermlineFilter | Germline leakage detection | Identifies germline variants misclassified as somatic [39] | |
| omnomicsQ | Real-time sequencing QC | Flags low-quality samples pre-analysis [42] | |
| Annotation & Interpretation | ANNOVAR | Functional variant annotation | Comprehensive gene-based, region-based annotations |
| Ensembl VEP | Variant effect prediction | Impact on genes, transcripts; plugin architecture [42] | |
| CIViC, COSMIC | Clinical evidence | Therapeutic, prognostic, diagnostic implications | |
| Machine Learning | XGBoost/LightGBM | Tumor-only classification | Reduces false positives; mitigates racial bias [38] |
| GradientBoosting | Confidence classification | Identifies high-confidence variants to reduce confirmatory testing [44] |
Regulatory and Quality Assurance Frameworks
For laboratories implementing variant calling pipelines for clinical or translational chemogenomic applications, adherence to established regulatory and quality assurance frameworks is essential:
- ISO 13485:2016: Defines quality management system requirements for medical devices and in vitro diagnostic products, ensuring documented design processes and risk management [42]
- IVDR (In Vitro Diagnostic Regulation): EU regulation requiring rigorous clinical evidence and performance evaluation for diagnostic tests [42]
- ACMG/AMP/ASCO/CAP Guidelines: Provide tiered systems for interpreting somatic variants based on clinical significance [42]
- External Quality Assessment (EQA): Participation in programs like EMQN and GenQA enables cross-laboratory benchmarking and performance validation [42]
The accurate discrimination between somatic and germline variants requires a multifaceted approach tailored to specific research contexts and available resources. For chemogenomic screens focused on drug response biomarkers, the following strategic recommendations emerge from current evidence:
- Implement ensemble approaches combining Mutect2, Strelka, and at least one additional caller to maximize both SNV and indel detection accuracy while maintaining computational efficiency [41].
- Adopt machine learning classifiers for tumor-only analyses to significantly reduce false positive rates and eliminate racial biases inherent in database-filtering approaches [38].
- Rigorously assess sample quality through TIN contamination evaluation, particularly for hematological malignancies and sarcomas where contamination prevalence is highest [40].
- Utilize systematic quality segmentation to identify the 10% of genomic regions responsible for the majority of false positives, enabling targeted filtering without excessive reduction in sensitivity [43].
- Validate findings in difficult genomic regions, particularly for pharmacogenes with high homology (e.g., CYP2D6, UGT2B17) where standard short-read technologies may underperform [37] [36].
As chemogenomic screens continue to evolve, integrating these robust variant calling strategies will ensure the reliability of drug-gene associations and accelerate the development of personalized cancer therapies.
In the rigorous field of chemogenomic screens, where the identification of genetic variants underpins critical discoveries in drug target identification and mechanism of action studies, the precision of next-generation sequencing (NGS) data analysis is paramount. The pathway from raw sequencing reads to confident variant calls is a complex computational process, with pre-processing steps forming the foundational layer upon which all subsequent analysis rests. This guide objectively examines the empirical evidence for three cornerstone pre-processing operations—read trimming, duplicate marking, and Base Quality Score Recalibration (BQSR)—in the context of accurate variant calling for drug discovery research. By comparing the performance of various tools and methodologies against benchmark datasets, we provide a scientific basis for optimizing NGS pipelines to enhance the reliability of variant data in chemogenomic applications.
The Foundation: Core Pre-processing Steps and Their Mechanisms
Read Trimming
Read trimming is a procedure intended to remove two types of potentially problematic sequences from raw NGS reads: low-quality bases, which cluster primarily at the 3' ends of reads, and residual adapter sequences. The theoretical basis for trimming is that excluding these artifacts should lead to more accurate read alignment and, consequently, more reliable variant calls [45]. Common tools for this task include Trimmomatic, fastp, BBduk, and Trim Galore, each employing distinct algorithms for adapter detection and quality-based trimming [45] [46].
Duplicate Marking
Duplicate marking involves identifying and flagging read pairs that appear to originate from the same original DNA fragment, most often a consequence of PCR amplification during library preparation. These duplicates can skew variant allele frequencies and create false positive calls. Tools like Picard Tools MarkDuplicates and Sambamba are routinely used to identify these redundant reads based on their alignment coordinates, ensuring that the downstream variant caller considers only unique evidence from the original DNA template [14] [8].
Base Quality Score Recalibration (BQSR)
BQSR is a sophisticated correction method implemented in pipelines such as the Genome Analysis Toolkit (GATK) Best Practices. It operates on the understanding that the base quality scores initially assigned by the sequencing instrument can be systematically inaccurate. BQSR builds an empirical error model by analyzing the data against known variant sites, then adjusts these quality scores to more accurately reflect the true probability of a base-calling error. This recalibrated data provides a more trustworthy input for the variant caller's probabilistic genotyping models [14] [8].
Tool Performance and Benchmarking Data
The following tables synthesize quantitative findings from systematic evaluations of pre-processing tools and their impact on variant calling accuracy.
Table 1: Impact of Read Trimming on Variant Calling Concordance in Bacterial Genomes
| Study Scope | Trimming Tools Tested | Key Finding (SNPs) | Key Finding (Indels) |
|---|---|---|---|
| >6,500 bacterial datasets (E. coli, M. tuberculosis, S. aureus) [45] | Atropos, fastp, Trim Galore, Trimmomatic | 98.8% of ~125 million SNPs were identically called with/without trimming | 91.9% of ~1.25 million indels were identically called with/without trimming |
| 17 Gram-negative bacterial genomes [45] | Atropos, fastp, Trim Galore, Trimmomatic | Minimal, statistically insignificant increase in SNP-calling accuracy | N/A |
Table 2: Impact of Adapter Trimming on Germline Variant Calling in the Human Genome
| Sequencing Type | Variant Callers Tested | Impact of Adapter Trimming | Notes |
|---|---|---|---|
| Whole-Genome Sequencing (WGS) [46] | DeepVariant, GATK, Clair3, Octopus, Strelka2, FreeBayes | No measurable effect on accuracy (Precision, Recall, F1 score) | Effect was negligible for best-performing callers (e.g., DeepVariant). |
| High-Coverage Whole-Exome Sequencing (WES) [46] | Multiple (see WGS) | Subtle improvement: 2-4 additional true positive variants in 2/7 samples. | Effect was not consistent and not dependent on adapter proportion. |
| Moderate-Coverage WES (~80-100x) [46] | Multiple (see WGS) | Negative impact on accuracy in some cases. | Suggests potential over-trimming with lower coverage data. |
Table 3: Performance of Popular Aligners and Variant Callers
| Tool Category | Tools | Performance Notes |
|---|---|---|
| Short Read Aligners [47] [17] | BWA-MEM, Novoalign, Bowtie2, Isaac | BWA-MEM is considered a gold standard. Bowtie2 performed significantly worse in one benchmark and is not recommended for medical variant calling [17]. |
| Variant Callers [48] [17] | DeepVariant, GATK, DRAGEN, Strelka2, Clair3, Octopus | DeepVariant consistently shows top performance and high robustness [17]. DRAGEN and DeepVariant show better accuracy than GATK, with no significant F1-score differences between them [48]. |
Experimental Protocols for Key Benchmarking Studies
To ensure reproducibility and provide a clear framework for evaluation, here are the detailed methodologies from several pivotal studies cited in this guide.
- Dataset: 17 sets of 150 bp Illumina HiSeq 4000 paired-end reads from Gram-negative bacteria and >6,500 publicly archived sequencing datasets.
- Trimming Tools & Parameters: Four trimmers (Atropos, fastp, Trim Galore, Trimmomatic) were used. The focus was on a "minimum-effort" application: automatic adapter detection (where available) and 3' quality-trimming across a range of stringencies.
- Alignment & Variant Calling: Reads (trimmed and untrimmed) were aligned with BWA-MEM. SNPs and indels were called using three variant callers: LoFreq, mpileup, and Strelka2.
- Validation: For the 17-genome set, a high-confidence truth set of SNPs was generated from whole-genome alignments of closed assemblies from both ONT and PacBio long reads.
- Dataset: 14 "gold standard" Genome-in-a-Bottle (GIAB) samples with established truth sets for both WGS and WES data.
- Trimming Tools & Parameters: fastp, Trimmomatic, and BBduk were used in palindromic mode with built-in Illumina adapter databases.
- Alignment & Pre-processing: All reads were aligned with BWA-MEM, and duplicates were marked with GATK MarkDuplicates.
- Variant Calling & Benchmarking: Six variant callers (Clair3, DeepVariant, Freebayes, GATK-HC, Octopus, Strelka2) were used. Performance was evaluated against the GIAB truth sets using the hap.py toolkit, with analysis restricted to coding sequences (CDS).
Visualizing the NGS Pre-processing and Variant Calling Workflow
The following diagram illustrates the standard workflow for NGS data pre-processing and variant calling, highlighting the three key steps examined in this guide.
Table 4: Key Resources for Robust NGS Pipeline Development
| Resource Name | Category | Primary Function in Validation |
|---|---|---|
| Genome in a Bottle (GIAB) [14] [46] [48] | Benchmark Dataset | Provides gold-standard, high-confidence variant calls for several human genomes, enabling objective accuracy measurement. |
| Platinum Genomes [48] | Benchmark Dataset | Another widely used set of benchmark variant calls for human samples, often used in conjunction with GIAB. |
| Synthetic Diploid (Syndip) [14] [48] | Benchmark Dataset | Derived from long-read assemblies of haploid cell lines, provides a less biased benchmark, especially in challenging genomic regions. |
| hap.py [46] [17] | Benchmarking Tool | A GA4GH-compliant software toolkit for the precise comparison of variant calls against a truth set, supporting stratified performance analysis. |
The imperative for meticulous pre-processing in NGS variant calling is nuanced. The collective evidence indicates that while duplicate marking remains a non-negotiable step to prevent amplification biases, the value of read trimming is highly context-dependent, offering negligible benefits for germline small variant calling with modern aligners and callers, particularly in WGS. BQSR continues to be recommended in best-practice pipelines, though its marginal gains must be weighed against its computational cost. For chemogenomic screens, where the accurate detection of true positive variants directly impacts the identification of viable drug targets, the selection of a variant caller has been shown to be more impactful than the choice of pre-processing tool. Researchers are advised to prioritize the implementation of robust, modern callers like DeepVariant or DRAGEN and to validate their entire pipeline, including pre-processing steps, against relevant benchmark resources like GIAB to ensure optimal accuracy for their specific experimental context.
In the field of chemogenomics, where high-throughput screening identifies interactions between chemical compounds and gene products, the accuracy of next-generation sequencing (NGS) variant calling is foundational to generating reliable biological insights. The choice between short-read and long-read sequencing technologies represents a critical methodological decision that directly impacts data quality, variant detection capability, and ultimately, the validity of research conclusions. While short-read sequencing has long served as the workhorse for NGS applications due to its high accuracy and cost-effectiveness, emerging long-read sequencing platforms now challenge this paradigm by offering unprecedented ability to resolve complex genomic regions and structural variations [49]. This guide provides an objective, data-driven comparison of these competing technologies within the specific context of chemogenomic screens, empowering researchers to make evidence-based decisions that optimize sequencing strategies for their specific research objectives.
The fundamental differences between these technologies are not merely technical but have profound implications for experimental outcomes. As recent benchmark studies demonstrate, platform-specific variations in sequencing depth, coverage uniformity, and error profiles can significantly affect the detection of genetic variants in chemogenomic fitness assays [50] [51]. By examining quantitative performance metrics across platforms and providing detailed experimental methodologies, this guide aims to equip researchers with the knowledge needed to navigate the complex landscape of modern sequencing technologies.
Short-Read Sequencing Technologies
Short-read sequencing technologies, often classified as second-generation sequencing, are characterized by their ability to sequence millions of DNA fragments in parallel through sequencing-by-synthesis or sequencing-by-ligation approaches. These platforms typically produce reads of 50-300 base pairs in length, which are then computationally assembled against a reference genome [52] [53]. The dominant short-read platform, Illumina, utilizes bridge amplification on flow cells to generate clustered DNA templates that are sequentially read using fluorescently-labeled nucleotides. Other notable short-read technologies include Element Biosciences' AVITI System, which employs sequencing-by-binding (SBB) chemistry for high accuracy (Q40+), and Thermo Fisher Scientific's Ion Torrent, which detects nucleotide incorporation through pH changes rather than optical signals [52] [53].
The key advantages of short-read technologies include their exceptionally high base-level accuracy (exceeding 99.9%), high throughput capabilities, and relatively low cost per base [54]. These characteristics have made short-read sequencing the preferred choice for applications requiring precise single-nucleotide variant calling, including chemogenomic screens that detect subtle fitness differences through barcode sequencing. However, a significant limitation of short-read technologies is their inability to resolve long repetitive regions or complex structural variations due to the fragmentation and amplification steps required for library preparation [52] [49]. This fragmentation process can introduce biases and makes it challenging to phase genetic variants or resolve complex genomic architectures.
Long-Read Sequencing Technologies
Long-read sequencing, often referred to as third-generation sequencing, encompasses technologies capable of sequencing individual DNA molecules without fragmentation, producing reads that typically range from 5,000 to over 100,000 base pairs [52] [55]. The two predominant long-read platforms are Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). PacBio's Single Molecule Real-Time (SMRT) sequencing immobilizes DNA polymerase at the bottom of nanoscale wells called zero-mode waveguides, monitoring nucleotide incorporation in real-time. The platform's HiFi (High Fidelity) mode, which uses circular consensus sequencing (CCS) to read DNA molecules multiple times, achieves accuracies exceeding 99.9% [52] [55]. Oxford Nanopore technologies employ a fundamentally different approach, threading single DNA strands through protein nanopores and detecting nucleotide-specific changes in ionic current [52].
The primary advantage of long-read technologies is their ability to span complex genomic regions, including repetitive elements, structural variants, and GC-rich sequences that are problematic for short-read platforms [55] [37]. This capability makes them particularly valuable for resolving complex pharmacogenes and structural variations relevant to chemogenomics. Additionally, long-read platforms can directly detect epigenetic modifications such as DNA methylation without specialized library preparation methods [49]. Historically, long-read technologies were limited by higher error rates and greater costs, but recent advancements have substantially improved their accuracy and affordability, making them increasingly competitive for large-scale genomic studies [55].
Table 1: Key Characteristics of Major Sequencing Platforms
| Platform | Technology Type | Read Length | Key Strengths | Primary Limitations |
|---|---|---|---|---|
| Illumina NovaSeq | Short-read | 50-300 bp | High accuracy (>99.9%), high throughput, low cost per base | Limited ability to resolve repeats and structural variants |
| Element Biosciences AVITI | Short-read | Up to 300 bp | Q40+ accuracy, flexible throughput | Similar limitations as other short-read platforms for complex genomics |
| PacBio Revio | Long-read (HiFi) | 10-25 kb | >99.9% accuracy, excellent for structural variants | Higher DNA input requirements, higher cost for some applications |
| Oxford Nanopore PromethION | Long-read | 5 bp - >1 Mb | Ultra-long reads, direct epigenetic detection, portable | Higher raw error rate requires deeper coverage for consensus accuracy |
Performance Benchmarking: Experimental Data and Metrics
Sequencing Depth, Coverage, and Accuracy
Recent cross-platform benchmarking studies provide critical insights into the performance characteristics of short-read and long-read technologies for variant identification. A comprehensive 2025 study comparing Illumina NovaSeq, ONT MinION, and PacBio Sequel II for SARS-CoV-2 genomic surveillance revealed that NovaSeq produced the highest number of reads and bases, resulting in superior depth of coverage and more complete consensus genomes [50]. The long-read platforms demonstrated lower yields and sequencing depth, which initially limited their variant identification capabilities, though implementing proper quality controls achieved consistent lineage assignments across all platforms [50].
In a methodological comparison focused on colorectal cancer samples, researchers conducted a rigorous analysis of variant calling performance between Illumina and Nanopore technologies [54]. The study reported that Illumina sequencing achieved approximately 105X coverage across target regions, while Nanopore whole-genome sequencing attained mean coverage of 16-28X, reflecting the different depth requirements for accurate variant calling between platforms [54]. Despite these coverage differences, the study found that Nanopore sequencing demonstrated enhanced ability to resolve large and complex structural variants, with consistently high precision across variant classes.
Base-level accuracy metrics further illuminate platform-specific performance characteristics. The same colorectal cancer study analyzed mapping quality scores, finding that Illumina achieved a mapping accuracy of 99.96% compared to 99.89% for Nanopore [54]. While this difference appears modest, it can have significant implications for detecting low-frequency variants in heterogeneous samples, a common challenge in chemogenomic screens assessing population-level fitness effects.
Optimal Sequencing Depth for Barcode-Based Screens
Chemogenomic fitness screens frequently rely on sequencing DNA barcodes to quantify the relative abundance of different mutants under chemical perturbation. Determining the optimal sequencing depth for these experiments is crucial for maximizing data quality while conserving resources. A 2025 study specifically addressed this challenge by analyzing noise characteristics in NGS counts from barcoded libraries [56].
Contrary to conventional wisdom, this research demonstrated that increasing sequencing depth does not always improve measurement precision for barcode concentration quantification. The study found that noise in NGS counts increases with sequencing depth, creating a point of diminishing returns beyond which deeper sequencing fails to enhance data quality [56]. Through mathematical modeling that accounted for PCR amplification biases in library preparation, the authors proposed that the optimal sequencing depth should be approximately ten times the initial amount of barcoded DNA molecules before any amplification step [56].
This finding has profound implications for designing efficient and cost-effective chemogenomic screens. Rather than simply maximizing sequencing depth, researchers should carefully estimate their library complexity and apply this rule of thumb to determine the appropriate depth for their specific experiment, potentially realizing significant cost savings without compromising data quality.
Table 2: Performance Metrics from Comparative Sequencing Studies
| Performance Metric | Illmina NovaSeq | PacBio Sequel II | ONT MinION |
|---|---|---|---|
| Relative Read Yield | Highest | Lower | Lower |
| Coverage Stability | Most stable across ORFs | Variable | Variable |
| Consensus Genome Completeness | Highest | Quality-dependent | Quality-dependent |
| Variant Calling Accuracy | >99.9% | >99.9% (HiFi mode) | ~99% |
| Structural Variant Detection | Limited | Excellent | Excellent |
| Typical Coverage for Variant Calling | 100-150X | 20-30X (HiFi) | 30-50X |
Experimental Protocols for Cross-Platform Comparison
Library Preparation Methodologies
Standardized library preparation is essential for meaningful cross-platform performance comparisons. For short-read Illumina sequencing, the typical workflow begins with cDNA synthesis using the SuperScript IV first-strand synthesis system with thermal cycler conditions of 25°C for 10 minutes, 50°C for 30 minutes, and 80°C for 10 minutes [50]. Libraries are then constructed using amplicon-based kits such as IDT's xGen Amplicon Core Kit for SARS-CoV-2, following manufacturer instructions for normalization and pooling. The final pool is sequenced on an Illumina NovaSeq using appropriate reagent kits [50].
For Oxford Nanopore long-read sequencing, library preparation typically involves reverse transcription followed by PCR amplification with kits such as the Midnight RT PCR Expansion. The reverse transcription reaction is performed at 25°C for 2 minutes, 55°C for 10 minutes, and 95°C for 1 minute [50]. PCR amplification then proceeds with initial denaturation at 95°C for 30 seconds, followed by 35 cycles of denaturation at 98°C for 15 seconds, and annealing/extension at 61°C for 2 minutes and 65°C for 3 minutes. Primer pools are combined, and libraries are prepared using rapid barcoding kits before sequencing on MinION flow cells [50].
Pacific Biosciences long-read sequencing employs distinct library preparation methods, beginning with cDNA transformation using the Molecular Loop Viral RNA Capture Kit with thermal cycler conditions of 25°C for 10 minutes, 50°C for 50 minutes, and 95°C for 1 minute, followed by 24-hour probe hybridization at 55°C [50]. Samples are barcoded using M13 barcodes and amplified with 26 cycles of 95°C for 3 minutes, 98°C for 15 seconds, 55°C for 15 seconds, and 72°C for 90 seconds. The barcoded cDNA samples are pooled and purified before library preparation with the SMRTbell Express Template prep kit, followed by sequencing on the Sequel II instrument [50].
Quality Control and Bioinformatics Processing
Robust quality control and standardized bioinformatics processing are crucial for ensuring comparable results across sequencing platforms. For all platforms, raw sequencing data in FASTQ format should undergo trimming and quality filtering using tools like fastp with standardized parameters: qualifiedqualityphred:15; unqualifiedpercentlimit:40; nbaselimit:5; complexitythreshold:30; polygminlen:10; polyxminlen:10; cutmeanquality:20; and overrepresentationsampling:20 [50]. All reads should be filtered with a minimum length of 40 bp, and FASTQ quality should be assessed with FastQC, with reports aggregated using MultiQC.
For read alignment, researchers should use platform-specific mapping settings in minimap2: -x sr for Illumina NovaSeq, map-ont for ONT MinION, and map-pb for PacBio Sequel II reads [50]. The resulting SAM files should be converted to sorted BAM files using Samtools, followed by variant calling with bcftools. Consensus genomes can be generated using iterative refinement approaches such as IRMA (Iterative Refinement Meta-Assembler) with appropriate modules and parameters [50].
Specific quality thresholds should be established for including sequencing data in downstream analyses. Based on recent benchmarking studies, recommended passing metrics include: percent ambiguous nucleotides (N) < 10% and reference genome coverage > 90% [50]. For phylogenetic analysis and lineage assignment, consensus sequences should be aligned against reference genomes using MAFFT followed by phylogenetic placement with FastTree [50].
Diagram 1: Comparative sequencing workflow for short-read and long-read platforms
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of sequencing-based chemogenomic screens requires access to specialized reagents and computational tools. The following table summarizes essential resources referenced in recent comparative studies.
Table 3: Essential Research Reagents and Computational Tools for Sequencing Studies
| Category | Specific Product/Software | Application Purpose | Key Features/Benefits |
|---|---|---|---|
| Library Prep Kits | IDT xGen Amplicon Core Kit | Illumina short-read library preparation | Optimized for target enrichment, uniform coverage |
| ONT Midnight RT PCR Expansion | Nanopore long-read library prep | Designed for complete genome amplification in few reactions | |
| PacBio SMRTbell Express Template Prep Kit | PacBio long-read library preparation | Optimized for structural variant detection and haplotype phasing | |
| Enzymes & Reagents | SuperScript IV First-Strand Synthesis System | cDNA synthesis for RNA viruses | High thermostability and processivity |
| PrimeSTAR GXL DNA Polymerase | High-fidelity PCR amplification | Excellent accuracy for amplicon generation | |
| Sequencing Platforms | Illumina NovaSeq 6000 | High-throughput short-read sequencing | Highest output for population-level studies |
| Oxford Nanopore MinION | Portable long-read sequencing | Real-time analysis, adjustable read length | |
| PacBio Sequel II | High-accuracy long-read sequencing | HiFi reads for superior variant calling | |
| Bioinformatics Tools | fastp (v0.23.4) | Quality control and adapter trimming | Rapid processing with integrated quality reporting |
| minimap2 (v2.17) | Sequence alignment to reference | Platform-specific presets for optimal mapping | |
| bcftools (v1.10.2) | Variant calling and manipulation | Flexible variant detection from BAM files | |
| IRMA (v1.1.3) | Iterative consensus generation | Modular design for different pathogen types |
Strategic Implementation in Chemogenomic Research
Platform Selection Guidelines for Specific Applications
The optimal sequencing platform choice for chemogenomic research depends heavily on the specific experimental goals and genomic targets. For large-scale barcode-based fitness screens that quantify mutant abundance through short DNA barcodes, short-read sequencing platforms typically provide the most cost-effective solution [56] [51]. The high accuracy and throughput of Illumina platforms make them ideal for detecting subtle abundance differences in pooled screens, provided sequencing depth is optimized according to library complexity (approximately 10X the initial barcode molecule count) [56].
For chemogenomic studies focusing on complex genomic regions, including those with repetitive elements, structural variations, or high GC content, long-read technologies offer distinct advantages. Research on pharmacogenetically important genes such as CYP2D6, CYP2B6, and HLA-B has demonstrated that long-read sequencing can resolve complex diplotypes and structural variants that are frequently mischaracterized by short-read approaches [37]. These capabilities make long-read sequencing particularly valuable for comprehensive variant annotation in genes with clinical relevance to drug response.
In studies requiring epigenetic profiling alongside variant detection, Oxford Nanopore technologies provide the unique ability to directly detect DNA methylation and other base modifications without specialized library preparation [55] [49]. This integrated approach can reveal relationships between genetic variation and epigenetic regulation in chemical response, offering a more comprehensive view of compound mechanisms of action.
Integrated Approaches and Future Directions
Rather than viewing short-read and long-read technologies as mutually exclusive, forward-looking chemogenomic research is increasingly adopting integrated sequencing strategies that leverage the complementary strengths of both platforms [49]. A hybrid approach utilizes short-read data as a scaffolding foundation for high-confidence single-nucleotide variant calling, while incorporating long-read data to resolve complex structural variations and phase haplotypes [55] [49]. This strategy proves particularly powerful for de novo assembly of microbial genomes or for characterizing complex eukaryotic genomes with high repetitive content.
The sequencing technology landscape continues to evolve rapidly, with both short-read and long-read platforms demonstrating substantial improvements in accuracy, throughput, and cost-effectiveness. Recent entrants to the short-read market, including Element Biosciences and Ultima Genomics, are driving increased competition and innovation [52] [53]. Simultaneously, PacBio's Revio system and Oxford Nanopore's PromethION platforms are making long-read sequencing more accessible for large-scale studies [52] [55]. For chemogenomic researchers, this dynamic environment necessitates ongoing evaluation of sequencing strategies to ensure methodological approaches remain aligned with technological capabilities.
Diagram 2: Decision framework for sequencing platform selection
The choice between short-read and emerging long-read sequencing technologies represents a fundamental strategic decision that directly impacts the quality and scope of variant calling in chemogenomic research. As comparative studies demonstrate, each platform offers distinct advantages: short-read technologies provide unparalleled accuracy and cost-efficiency for detecting single-nucleotide variants and quantifying barcode abundances, while long-read platforms excel at resolving complex genomic architectures, structural variations, and epigenetic modifications [50] [54] [55]. Rather than adhering to a one-size-fits-all approach, researchers should select sequencing strategies based on their specific experimental goals, target genomic features, and analytical requirements.
The evolving sequencing landscape offers exciting opportunities for methodological innovation in chemogenomics. By understanding the performance characteristics, optimal applications, and limitations of each platform, researchers can design more robust and informative studies. Furthermore, integrated approaches that combine both short-read and long-read data are increasingly feasible and offer the most comprehensive solution for challenging genomic targets [49]. As sequencing technologies continue to advance, maintaining awareness of platform capabilities will remain essential for generating reliable, actionable insights from chemogenomic screens.
Beyond the Basics: Advanced Strategies to Refine and Optimize Variant Detection
Tackling Tumor Heterogeneity and Low-Frequency Variants in Compound-Treated Pools
In modern oncology drug development, chemogenomic screens using compound-treated pools are essential for identifying candidate therapies. However, the accurate detection of low-frequency variants within these pools is severely complicated by tumor heterogeneity [57]. Tumor heterogeneity, a fundamental characteristic of malignant cancers, results in subpopulations of cells with diverse molecular profiles, leading to variations in growth rates, metastatic potential, and drug sensitivity [57] [58]. In the context of compound-treated pools, this heterogeneity means that drug-sensitive clones may be eliminated while resistant subclones survive and expand, often present initially as low-frequency variants undetectable by conventional sequencing methods.
The critical challenge lies in distinguishing genuine low-frequency somatic variants from sequencing artifacts and background noise, especially after compound exposure which alters clonal population dynamics. Next-generation sequencing (NGS) technologies provide the foundation for variant calling in these experiments, but each platform presents specific advantages and limitations for this application [59]. The accuracy of NGS variant calling directly impacts the reliability of chemogenomic screen outcomes, potentially determining whether promising drug candidates are identified or overlooked. This guide objectively compares the performance of current NGS-based technologies specifically for tackling tumor heterogeneity and detecting low-frequency variants in compound-treated pool scenarios, providing researchers with experimental data and methodologies to inform their technology selection.
Understanding Tumor Heterogeneity in Drug-Treated Pools
Origins and Impact of Tumor Heterogeneity
Tumor heterogeneity develops through multiple mechanisms that create diverse subclonal populations within tumors. Genomic instability serves as a primary driver, resulting from exposure to exogenous mutagenic sources or endogenous processes like DNA replication errors and oxidative stress [57]. This instability generates complex chromosomal rearrangements including gene losses, amplifications, and translocations that establish the genetic diversity upon which selection pressures can act.
The clonal evolution model provides a framework for understanding how heterogeneity influences treatment outcomes. In this model, cancer cells continuously acquire mutations, with selective pressures (such as anticancer drugs) promoting the expansion of resistant subclones [57]. This dynamic is particularly relevant in compound-treated pools, where pre-existing resistant variants may be present at frequencies below conventional detection limits but expand significantly under therapeutic pressure. Research has demonstrated that EGFR-mutant NSCLC tumors treated with tyrosine kinase inhibitors (TKIs) show clear evidence of this phenomenon, with sensitive clones diminishing while resistant clones progressively dominate [57].
Detection Challenges in Compound-Treated Pools
The accurate detection of low-frequency variants in compound-treated pools presents distinct technical challenges:
Variant Allele Frequency Thresholds: Conventional NGS variant calling typically requires variant allele frequencies (VAF) of 2-5% for reliable detection, potentially missing clinically relevant resistant subclones present at lower frequencies [57].
Spatial and Temporal Heterogeneity: Studies comparing multiple regions of the same tumor have revealed striking genetic diversity, with one kidney cancer study finding only 34% of mutations present across all sampled regions [57]. This spatial variation complicates predicting treatment response from limited sampling.
Dynamic Clonal Evolution: Continuous genomic changes during treatment create temporal heterogeneity, necessitating repeated monitoring to capture evolving resistance patterns [57].
Table 1: Key Challenges in Detecting Low-Frequency Variants in Compound-Treated Pools
| Challenge | Impact on Variant Detection | Consequence for Drug Screening |
|---|---|---|
| Pre-existing resistant subclones | Low initial VAF (<1%) | False negative calls for resistant variants |
| Clonal expansion under selection | Dynamic VAF changes | Inaccurate assessment of compound efficacy |
| Tumor mutational burden | High background variant noise | Reduced sensitivity for driver mutations |
| Technical artifacts | False positive variants | Misidentification of resistance mechanisms |
Comparative Analysis of NGS Technologies for Variant Detection
Technology Platforms and Performance Characteristics
Multiple sequencing technologies offer distinct approaches to addressing the challenge of low-frequency variant detection in heterogeneous samples. Each platform balances read length, accuracy, throughput, and cost differently, resulting in varied performance characteristics for specific applications in chemogenomic screens.
Short-read sequencing technologies (e.g., Illumina) remain the workhorse for most variant calling applications due to their high base-level accuracy (exceeding 99.9%) and tremendous throughput [60]. However, their limited read length (typically 75-300 bp) presents challenges in resolving complex genomic regions, structural variants, and phasing haplotypes - all critical for fully characterizing heterogeneous tumor samples [61].
HiFi (High Fidelity) sequencing from PacBio represents a significant advancement, combining long reads (10-25 kb) with high accuracy (>99.9%) through circular consensus sequencing (CCS) [61]. This technology provides more complete genomic context for variant calling, enabling researchers to distinguish between true low-frequency variants and technical artifacts with greater confidence. The long-range information allows for more accurate phasing of mutations, determining whether variants occur on the same or different DNA molecules - crucial information for understanding clonal architecture in heterogeneous pools.
Third-generation sequencing platforms like Oxford Nanopore offer ultra-long reads that can span complex genomic regions but have historically faced challenges with higher error rates that complicate low-frequency variant detection, though continuous improvements are addressing these limitations.
Table 2: Technology Comparison for Low-Frequency Variant Detection in Heterogeneous Pools
| Technology | Optimal VAF Detection Limit | Variant Type Strengths | Limitations for Heterogeneous Pools |
|---|---|---|---|
| Short-read WGS (Illumina) | 1-2% | SNVs, small indels | Limited phasing information, poor structural variant detection |
| Targeted NGS | 0.5-1% | Predefined SNVs/indels | Restricted to panel regions, may miss novel variants |
| HiFi Sequencing (PacBio) | 1-2% | Structural variants, phased SNVs | Higher DNA input requirements, cost considerations |
| ddPCR | 0.1-0.001% | Ultra-sensitive for known mutations | Limited to few predefined mutations, not discovery-based |
Experimental Performance Data in Controlled Studies
Comparative studies provide quantitative performance data for different technologies in detecting low-frequency variants. In one comprehensive analysis, targeted NGS approaches demonstrated reliable detection of variants at allele frequencies as low as 0.5-1% with sufficient coverage depth (>500x), making them suitable for monitoring known resistance mutations in compound-treated pools [59].
Research specifically evaluating HiFi sequencing for variant calling revealed its particular strength in detecting structural variants (SVs) and insertion-deletion events (indels) that are frequently missed by short-read technologies [61]. In the HG002 reference genome study, HiFi reads detected SVs with an F1 score of 92.5%, significantly outperforming short-read technologies for this variant class [61]. This capability is crucial for comprehensive variant profiling in heterogeneous samples where structural variants may drive resistance.
For the most sensitive detection of known variants at very low frequencies (<0.1%), digital PCR (dPCR) remains the gold standard, capable of quantitative analysis of mutation frequencies as low as 0.001%-0.0001% [57]. However, this extreme sensitivity comes at the cost of limited multiplexing capability and inability to discover novel variants.
Experimental Protocols for Technology Validation
Cell Line Mixing Studies for Sensitivity Determination
A robust approach for determining the sensitivity limits of variant calling in compound-treated pools involves creating defined mixtures of cell lines with known genomic profiles:
Protocol:
- Cell Line Selection: Identify two or more cancer cell lines with comprehensively characterized mutational profiles, including specific single nucleotide variants (SNVs), indels, and copy number alterations.
- Sample Preparation: Mix genomic DNA from these cell lines in precisely defined ratios (e.g., 50:50, 10:90, 1:99, 0.1:99.9) to simulate heterogeneous samples with known variant allele frequencies.
- Library Preparation: Process mixed samples using standardized library preparation protocols for each technology being evaluated (short-read WGS, targeted NGS, HiFi sequencing).
- Sequencing and Analysis: Sequence all libraries to appropriate depth and process through identical bioinformatic pipelines for variant calling.
- Sensitivity Calculation: Calculate sensitivity as (Number of variants detected at known positions / Total number of known variants) × 100% for each mixing ratio and technology.
This controlled experimental design enables direct comparison of the limit of detection (LOD) for each technology and reveals technology-specific biases in variant calling.
Compound-Treated Pool Monitoring Workflow
For evaluating technology performance in actual drug discovery contexts, implement a longitudinal monitoring protocol:
Protocol:
- Pool Generation: Create a diverse cell pool by combining multiple cancer cell lines or using patient-derived organoids with inherent heterogeneity.
- Compound Treatment: Expose pools to candidate compounds at relevant concentrations, including appropriate controls (DMSO vehicle, etc.).
- Timepoint Sampling: Collect samples at multiple timepoints (e.g., pre-treatment, 24h, 72h, 1-week) to capture dynamic clonal changes.
- Multi-technology Sequencing: Process identical samples across all comparison technologies in parallel.
- Variant Calling and Clonal Tracking: Implement appropriate variant calling pipelines for each technology and track specific variants across timepoints.
This workflow generates comparative data on each technology's ability to detect emergent resistance variants and track clonal dynamics in response to compound treatment.
Diagram 1: Experimental workflow for technology comparison in compound-treated pools
Essential Research Reagents and Solutions
Successful detection of low-frequency variants in heterogeneous samples requires careful selection and implementation of research reagents and solutions throughout the experimental workflow.
Table 3: Essential Research Reagent Solutions for Low-Frequency Variant Detection
| Reagent Category | Specific Examples | Function in Workflow | Performance Considerations |
|---|---|---|---|
| DNA Extraction Kits | QIAamp DNA Mini Kit, DNeasy Blood & Tissue Kit | High-quality, high-molecular-weight DNA extraction | Integrity critical for long-read technologies; minimize shearing |
| Library Preparation | Illumina DNA Prep, PacBio SMRTbell Prep, Swift Accel-NGS | Fragment processing and adapter ligation | Optimization needed for input amount and fragment size distribution |
| Target Enrichment | IDT xGen Panels, Twist Human Core Exome | Selective capture of genomic regions | Efficiency impacts uniformity and off-target rates |
| QC & Quantification | Qubit dsDNA HS Assay, Agilent TapeStation, Fragment Analyzer | Quality assessment pre-sequencing | Critical for accurate library pooling and loading calculations |
| Unique Molecular Identifiers | IDT UMI Adapters, Twist UMI Kit | Tagging original molecules to correct PCR errors | Essential for distinguishing true low-frequency variants from artifacts |
| Variant Calling Pipelines | GATK, DeepVariant, Longshot | Bioinformatics analysis of sequencing data | Algorithm selection significantly impacts sensitivity/specificity balance |
Bioinformatics Considerations for Accurate Variant Calling
Specialized Analysis Pipelines
The accurate identification of low-frequency variants in heterogeneous samples requires specialized bioinformatic approaches beyond standard variant calling pipelines. Unique Molecular Identifiers (UMIs) have become essential tools, enabling bioinformatic correction of PCR amplification errors and sequencing artifacts by tagging original DNA molecules before amplification [62]. Implementation of UMI-based error correction can improve detection sensitivity by 10-100 fold for variants in the 0.1-1% VAF range.
For tumor heterogeneity analysis, several computational methods have been specifically developed to deconvolve mixed cell populations and infer clonal architecture from bulk sequencing data. Tools like PyClone, SciClone, and EXPANDS employ Bayesian clustering approaches to group mutations into putative clones based on their variant allele frequencies and copy number profiles. When applied to compound-treated pools, these methods can track the rise and fall of specific clones in response to treatment, providing insights beyond simple variant frequency changes.
Data Management and Storage Requirements
The scale of data generated by comprehensive variant calling studies necessitates careful data management planning. According to expert consensus on NGS bioinformatics platforms, storage architecture must accommodate both immediate analysis needs and long-term archival requirements [62]. Specifically:
- Active Analysis Storage: High-performance storage systems with rapid access capabilities, typically configured in RAID arrays or network-attached storage (NAS) solutions.
- Backup Strategy: Implementation of the "Grandfather-Father-Son" backup approach, with real-time backup of sequencing raw data, weekly backup of analysis results, and monthly archival of processed datasets [62].
- Retention Policy: Raw sequencing data should be retained for at least one year actively, with backups maintained for 15 years, while analysis results require permanent archival in most research contexts [62].
Diagram 2: Bioinformatics workflow and data management for heterogeneity analysis
The accurate detection of low-frequency variants in compound-treated pools requires careful technology selection based on specific research objectives and constraints. Short-read targeted NGS provides the most practical solution for focused screening of known resistance mutations with moderate sensitivity (0.5-1% VAF) and relatively low cost [59]. For discovery-oriented studies where novel or structural variants may contribute to resistance, HiFi sequencing offers superior capability despite higher per-sample costs and more demanding DNA input requirements [61].
For the most challenging detection scenarios requiring ultimate sensitivity for known variants, a tiered approach combining technologies provides the optimal strategy: using HiFi or short-read WGS for comprehensive variant discovery, followed by highly sensitive dPCR or targeted NGS for specific variant monitoring across multiple timepoints in compound treatment studies.
As sequencing technologies continue to evolve, with single-cell sequencing and spatial transcriptomics emerging as powerful tools for resolving tumor heterogeneity, the capabilities for detecting and tracking low-frequency variants in complex pools will continue to improve [58]. Nevertheless, the fundamental principles of rigorous experimental design, appropriate controls, and validated bioinformatic analysis will remain essential for generating reliable data from chemogenomic screens regardless of the specific technology platform employed.
The accurate identification of genetic variants, including single nucleotide variants (SNVs) and insertions/deletions (indels), from next-generation sequencing (NGS) data is a foundational task in genomics research and clinical diagnostics. For chemogenomic CRISPR screens—where the goal is to understand gene-drug interactions on a massive scale—variant calling accuracy is particularly crucial, as it enables researchers to identify genetic modifiers of drug response. Traditional variant callers, such as the Genome Analysis Toolkit (GATK), rely on statistical models hand-tuned by experts. However, the emergence of artificial intelligence (AI) and machine learning (ML) has introduced a paradigm shift, reframing variant calling as an image classification problem to achieve unprecedented accuracy.
AI-based tools like DeepVariant (developed by Google) leverage deep learning to analyze sequencing data, demonstrating superior performance in benchmark studies across diverse sequencing platforms and sample types. This evolution is critical for chemogenomic research, where the genetic landscape of cell lines used in CRISPR screens must be accurately characterized to avoid confounding results. This guide provides an objective comparison of leading variant callers, with a focus on AI-driven tools, their performance metrics, and their specific applicability to the rigorous demands of chemogenomic screen analysis.
Performance Comparison of Major Variant Callers
Independent benchmarking studies consistently reveal that AI-based variant callers, particularly DeepVariant, outperform conventional tools in key accuracy metrics. The following tables summarize comparative data for SNV and indel calling across different sequencing technologies.
Table 1: SNV Calling Performance Across Different Sequencing Platforms (F1-Score %)
| Variant Caller | Type | Illumina WES | PacBio HiFi | ONT |
|---|---|---|---|---|
| DeepVariant | AI-based | 98.95% [63] | >99.9% [63] | 97.07% [63] |
| GATK | Conventional | ~97% [64] | ~99% [63] | N/A |
| DNAscope | AI-based | 94.48% [63] | >99.9% [63] | N/A |
| BCFTools | Conventional | 98.83% [63] | ~99% [63] | <90% [63] |
Table 2: Indel Calling Performance Across Different Sequencing Platforms (F1-Score %)
| Variant Caller | Type | Illumina WES | PacBio HiFi | ONT |
|---|---|---|---|---|
| DeepVariant | AI-based | 81.41% [63] | >99.5% [63] | 80.40% [63] |
| GATK | Conventional | ~80% (inferred) | <85% [63] | N/A |
| DNAscope | AI-based | 57.53% [63] | >99.5% [63] | N/A |
| BCFTools | Conventional | 81.21% [63] | <85% [63] | 0% [63] |
Beyond raw accuracy, other critical differentiators include:
- Mendelian Consistency: In trio-based studies (a gold standard for germline accuracy), DeepVariant demonstrated a significantly lower Mendelian error rate (3.09%) compared to GATK (5.25%), indicating better real-world performance in family studies [64].
- Computational Efficiency: BCFools and Platypus are generally the fastest tools, while GATK is among the slowest. DeepVariant's runtime is moderate, but it can require significant memory resources, especially for long-read data [63].
Experimental Protocols for Benchmarking
The superior performance of AI-based tools is validated through rigorous benchmarking protocols. Understanding these methodologies is key to interpreting the data and designing robust chemogenomic studies.
Standard Benchmarking with Gold-Standard Datasets
The most reliable performance data comes from studies using the Genome in a Bottle (GIAB) consortium's gold-standard reference samples (e.g., HG001, HG002) [18]. The typical workflow is as follows:
- Data Acquisition: Publicly available whole-genome or whole-exome sequencing data from GIAB samples are downloaded from sources like the NCBI Sequence Read Archive (SRA). These datasets are often derived from multiple platforms (Illumina, PacBio HiFi, Oxford Nanopore) [18] [63].
- Variant Calling: The same set of sequencing data is processed through each variant calling tool (e.g., DeepVariant, GATK, DNAscope, BCFTools) using default parameters and a common reference genome (GRCh38).
- Benchmarking Analysis: The output VCF files from each tool are compared against the GIAB high-confidence truth sets using specialized assessment tools like hap.py or the Variant Calling Assessment Tool (VCAT). These tools calculate key metrics [18]:
- Precision: The proportion of called variants that are true variants (1 - False Positive Rate).
- Recall (Sensitivity): The proportion of true variants that are successfully detected by the caller.
- F1-Score: The harmonic mean of precision and recall, providing a single metric for accuracy.
Figure 1: Variant Caller Benchmarking Workflow
Specialized Validation in Clinical and Research Contexts
For clinical translation, performance is often assessed in more realistic scenarios:
- Trio Concordance: Sequencing data from parent-child trios are used to calculate the Mendelian error rate—the proportion of genotype calls in the child that are inconsistent with the parental genotypes. A lower rate indicates higher accuracy [64].
- Diagnostic Yield: In clinical studies, the number of confirmed disease-causing variants detected by each pipeline is compared. For example, one study found DeepVariant detected 62 out of 63 known pathological variants, while GATK detected 61 [64].
The AI Advantage: Core Methodologies of DeepVariant and DeepSomatic
The performance gains of AI-based tools stem from their unique approach to variant calling. Unlike conventional tools that apply statistical models, DeepVariant transforms the problem into an image classification task.
- Candidate Identification: The tool first scans aligned sequencing reads (BAM/CRAM files) to identify genomic positions that may contain a variant.
- Pileup Image Generation: For each candidate position, it creates a multi-channel "pileup image." This image tensor represents the aligned sequencing reads around the candidate locus, with different channels encoding key information such as:
- Base call (e.g., A, C, G, T)
- Base quality score
- Mapping quality
- Read strand
- Support for alternative alleles
- Variant Classification: A convolutional neural network (CNN), based on an architecture like Inception-v3 or MobileNetV2, analyzes the pileup image. The CNN is trained on millions of examples to classify the locus as homozygous reference, heterozygous variant, or homozygous alternate [65] [66].
This method allows the AI to learn complex, non-linear patterns from the data that are difficult to encapsulate in hand-written statistical models, making it more robust to sequencing errors and artifacts.
For cancer research, the DeepSomatic tool extends this pipeline to analyze matched tumor-normal sample pairs. It generates pileup images from both tissues and uses a specialized model to classify variants as somatic mutations, germline variants, or technical artifacts. This is particularly valuable for chemogenomic screens aiming to identify synthetic lethal interactions or drug-resistance mechanisms. DeepSomatic has demonstrated an F1-score of 98.3% for SNPs on Illumina data, significantly outperforming tools like MuTect2 and Strelka2 [67].
Figure 2: DeepVariant's AI Calling Process
Application in Chemogenomic CRISPR Screens
In chemogenomic CRISPR screens, researchers use genome-wide CRISPR libraries to knock out genes in pooled cell populations, which are then treated with various chemical compounds (drugs). The goal is to identify genes whose loss confers resistance or sensitivity to a drug. Accurate variant calling is critical at two stages:
- Genomic Quality Control of Cell Lines: Ensuring the genetic background of the cell line used (e.g., RPE1-hTERT p53−/−) is well-characterized and free of confounding variants [68].
- Validation of Engineered Mutations: Verifying CRISPR-induced mutations in selected genes or pathways following the screen.
The high accuracy of DeepVariant, especially for indels, is a significant asset. CRISPR-Cas9 editing predominantly generates indel mutations, and accurately calling these is necessary for validating gene knockout. Furthermore, AI models like DeepChem-Variant—a modular, open-source framework integrating DeepVariant—have been tested specifically for CRISPR off-target detection, showing high sensitivity (79-92%) in recovering variants called by the standard DeepVariant pipeline [65].
Table 3: Essential Research Reagents and Tools for Chemogenomic Screens
| Item | Function/Description | Example/Source |
|---|---|---|
| CRISPR Library | A pooled collection of sgRNAs for genome-wide knockout screens. | TKOv3 library (70,948 sgRNAs targeting 18,053 genes) [68]. |
| Cell Line | An immortalized cell line suitable for high-throughput screening. | RPE1-hTERT p53−/− Flag-Cas9 [68]. |
| Selection Agent | Antibiotic for selecting successfully transduced cells. | Puromycin [68]. |
| Transduction Enhancer | Chemical to improve lentiviral infection efficiency. | Polybrene or Protamine Sulfate [68]. |
| Analysis Software | Computational tool for identifying hit genes from screen data. | MAGeCK-VISPR [69]. |
The integration of AI and machine learning into variant calling has unequivocally raised the standard for accuracy in genomic analysis. Tools like DeepVariant consistently demonstrate superior performance, especially for challenging indel calls and across diverse sequencing platforms. For researchers conducting chemogenomic CRISPR screens, adopting these AI-powered tools provides more reliable characterization of cellular models and validation of genetic edits, thereby reducing false positives and increasing confidence in the identification of gene-drug interactions.
The field continues to evolve rapidly, with tools like DeepSomatic extending these capabilities to somatic mutation detection in cancer. The trend is towards more specialized, yet user-friendly, AI models that can be integrated into scalable and reproducible bioinformatics pipelines, further empowering drug development professionals to extract robust biological insights from their complex genomic datasets.
Formalin-fixed, paraffin-embedded (FFPE) samples represent a cornerstone of clinical cancer research, with an estimated 50-80 million solid tumor specimens globally considered potentially suitable for next-generation sequencing (NGS) analysis [70]. These archives, often paired with detailed clinical annotations, provide an invaluable resource for biomarker discovery and chemogenomic screen validation. However, the very process that preserves tissue morphology—formalin fixation—simultaneously introduces extensive DNA damage that compromises genomic analysis. This creates a critical tension in molecular research: while fresh frozen (FF) samples remain the gold standard for nucleic acid integrity, FFPE specimens offer unparalleled access to vast clinical cohorts with long-term outcome data [71].
The implications for chemogenomic screening are profound. Accurate variant calling is foundational to identifying genetic determinants of drug response, yet FFPE-derived artifacts can generate false positives that mislead therapeutic predictions. Concurrently, batch effects introduced through variable sample processing can confound cross-study comparisons essential for validating findings. This guide systematically compares performance metrics between FFPE and alternative sample types, provides experimentally validated mitigation strategies, and delineates a framework for optimizing NGS experimental design to maximize variant calling accuracy.
Understanding FFPE-Induced Molecular Damage and Its Impact on NGS
Formalin fixation triggers multiple chemical processes that damage DNA and introduce sequencing artifacts. The predominant mechanisms include:
- Chemical Additions and Cross-links: Formaldehyde reacts with nucleophilic groups on DNA bases, creating modified bases with altered pairing capabilities and forming methylene bridges that create DNA-DNA and DNA-protein cross-links [70].
- Depurination and Fragmentation: Formalin fixation accelerates glycosidic bond cleavage, generating apurinic/apyrimidinic (AP) sites that lead to DNA backbone fragmentation into separate segments [70].
- Deamination: Spontaneous cytosine deamination to uracil results in C>T/G>A transitions, while deamination of 5-methylcytosine yields thymine [70].
The cumulative effect of these processes is both information loss (through fragmentation and polymerase blocking lesions) and false signal introduction (mainly through deamination events). The artifacts are not uniformly distributed across the genome but are magnified in AT-rich regions due to local strand separation creating a vicious cycle of further damage [70].
Figure 1: FFPE-induced DNA damage pathways and their impacts on NGS data quality.
Comparative Performance: FFPE Versus Fresh Frozen Samples
DNA Quality and Sequencing Metrics
Multiple studies have quantitatively compared NGS performance between matched FFPE-FF sample pairs, revealing consistent patterns of differential performance.
Table 1: Comparative sequencing metrics between FFPE and fresh frozen samples
| Metric | Fresh Frozen (Gold Standard) | FFPE Samples | Experimental Support |
|---|---|---|---|
| DNA Fragment Size | >1000 bp (high molecular weight) | 200-400 bp (highly fragmented) | Agarose gel electrophoresis; Bioanalyzer [72] |
| Chimeric Read Percentage | 0.26% | 0.51% (p<0.0001) | Large-scale WGS analysis of 11,014 FF vs 578 FFPE [73] |
| Insert Size | 477 bp | 391 bp (p<0.0001) | WGS analysis [73] |
| Mapping Rate | 94.1% | 93.4% (p<0.0001) | WGS analysis [73] |
| Coverage Uniformity | High uniformity | GC bias, AT depletion | Hybridization capture data [73] |
| C>T/G>A Artifact Rate | Baseline | 7-fold increase | Targeted sequencing of 13-year-old samples [70] |
| Indel Burden | Baseline | Order of magnitude increase | WGS analysis across multiple tumor types [73] |
Variant Concordance and Analytical Sensitivity
Despite quantitative differences in DNA quality, multiple studies demonstrate that optimized protocols can yield high variant concordance between FFPE and frozen samples.
Table 2: Variant calling performance comparison between sample types
| Performance Measure | Concordance Rate | Notes | Reference |
|---|---|---|---|
| SNV Concordance | >99.99% | Paired FFPE-frozen from 16 lung adenocarcinomas | [72] |
| SNV Detection Agreement | 96.8% | Same study comparing single nucleotide variants | [72] |
| Orthogonal Platform Agreement | >98% | Compared to SNP array genotyping | [72] |
| Actionable Variant Detection | No significant difference | Domain 1 variants in 168 actionable genes | [73] |
| Low VAF Variants (<10%) | Compromised | 7.7% of true PIK3CA/BRAF mutations would be lost with VAF filtering | [73] |
A critical finding from recent large-scale analyses is that traditional bioinformatic filtering of variants with VAF <10%—a common approach to minimize FFPE artifacts—inappropriately discards genuine clinically-actionable mutations. In one study, 7.7% of true PIK3CA and BRAF V600E mutations occurred at VAF <10% and would have been eliminated by such filtering [73].
Multidimensional Mitigation: Integrated Strategies Across the Experimental Workflow
Effective mitigation of FFPE artifacts requires a comprehensive approach spanning pre-analytical, analytical, and computational stages.
Pre-analytical and Wet-Lab Mitigation Strategies
Figure 2: Integrated multi-stage mitigation workflow for FFPE artifacts.
Alternative Fixation Protocols
Comparative studies of fixation methods demonstrate that alternatives to neutral buffered formalin (NBF) significantly reduce artifacts:
- Acid-Deprived Formalin (ADF) and Cold ADF: Showed significantly longer reads, lowest noise levels, and highest uniformity in targeted sequencing [74].
- Glyoxal Acid-Free (GAF): Performed intermediate between ADF and NBF but superior to traditional formalin [74].
- Impact on Mutational Signatures: NBF samples showed highest mutational signature 1 (aging and FFPE artifact related) at 37% versus 17% in coldADF samples [74].
DNA Extraction and Library Preparation
- DNA Repair Treatments: Pre-sequencing enzymatic repair of FFPE-DNA can address cross-links, abasic sites, and deaminated bases [70].
- Hybrid Capture vs. Amplicon Approaches: Hybrid capture methods tolerate more sequence mismatches and avoid allele dropout problems seen in amplification-based assays [75].
- PCR-Free Library Preparation: FFPE libraries undergoing PCR show indel burdens increased by an order of magnitude, suggesting PCR-free approaches reduce this specific artifact type [73].
Computational Mitigation and Batch Effect Correction
Batch effects represent a parallel challenge in multi-sample studies, particularly when integrating datasets from different processing batches, sequencing centers, or time periods.
Batch Effect Detection
Effective detection begins with quantifying batch effects through:
- Principal Component Analysis (PCA) of quality metrics: Clearly reveals batch effects not apparent in genotype-based PCA [76].
- Quality Metrics: Include percentage of variants confirmed in reference databases, transition/transversion ratios (expected 2.0-2.1 in genomic DNA), mean genotype quality, median read depth, and percent heterozygotes [76].
Batch Effect Correction Algorithms
Table 3: Computational methods for batch effect correction
| Method | Mechanism | Applicability | Implementation |
|---|---|---|---|
| ComBat | Empirical Bayes framework adjusting for mean and variance | Multiple data types (RNA-seq, radiomics, NGS metrics) | R package 'sva' [77] |
| Limma removeBatchEffect | Linear modeling with batch as covariate | Assumes additive batch effects | R package 'Limma' [77] |
| Harmony | Iterative clustering with batch integration | Single-cell and bulk sequencing | Python/R packages [78] |
| Mutual Nearest Neighbors (MNN) | Identifies mutual nearest neighbors across batches | Single-cell RNA sequencing | Various implementations [78] |
| Seurat Integration | Canonical correlation analysis and mutual nearest neighbors | Single-cell multimodal data | Seurat package [78] |
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 4: Key reagents and methods for FFPE NGS studies
| Reagent/Method | Function | Considerations for FFPE Samples |
|---|---|---|
| DNA Repair Enzymes | Repair cross-links, abasic sites, deaminated bases | Critical for highly degraded samples; multiple commercial kits available [70] |
| Hybrid Capture Probes | Target enrichment for sequencing | Prefer over amplicon approaches; tolerate mismatches better [75] |
| PCR-Free Library Prep Kits | Minimize PCR-introduced errors | Reduce indel artifacts; particularly valuable for FFPE [73] |
| Quality Control Assays | Assess DNA fragmentation and quality | DNA Integrity Number (DIN), DV200; multiplex PCR for GAPDH [72] |
| Reference Standards | Benchmark variant calling accuracy | Genome in a bottle; cell line mixtures with known variants [75] |
| Batch Correction Software | Remove technical variation | ComBat, Limma for bulk data; Harmony, Seurat for single-cell [78] [77] |
Experimental Design Recommendations for Chemogenomic Screens
Sample Selection and Quality Assessment
- Prioritize Sample Matches: When possible, utilize matched FFPE-FF pairs from the same tumor to enable cross-validation [72].
- Systematic Quality Control: Implement quantitative metrics including DNA integrity numbers, fragment size distribution, and pre-sequencing QC assays [70] [75].
- Pathological Review Essential: Mandate pathologist assessment of tumor cellularity and selection of non-necrotic regions for macrodissection [75].
Strategic Experimental Design
- Batch Balancing: Distribute samples from different experimental conditions (e.g., drug treatments) across processing batches [76].
- Reference Standards Integration: Include common reference samples across batches to monitor technical variability [75].
- Replication Strategy: Plan for technical replicates of a subset of samples to quantify batch effects [76].
Bioinformatic Pipeline Specifications
- FFPE-Specific Filtering: Implement artifact-aware filtering rather than simple VAF thresholds [73].
- Batch Effect Monitoring: Conduct PCA of quality metrics, not just genotypes, to detect technical artifacts [76].
- Mutational Signature Analysis: Characterize and quantify FFPE-specific signatures (SBS-FFPE, ID-FFPE) to measure artifact burden [73].
FFPE samples present both challenges and unparalleled opportunities for comprehensive chemogenomic analysis. While fresh frozen specimens remain the gold standard for DNA integrity, methodological advances across the entire workflow—from alternative fixation protocols to bioinformatic artifact characterization—now enable reliable variant calling from FFPE-derived DNA. The critical realization is that FFPE and fresh frozen samples are not interchangeable without appropriate adjustments, but with optimized protocols, FFPE specimens can yield clinically actionable data with >99.99% concordance for single nucleotide variants [72].
Successful integration of FFPE samples into chemogenomic screens requires a multidimensional approach: (1) implementing pre-analytical controls including consideration of alternative fixatives; (2) selecting appropriate wet-lab methods including hybrid capture and DNA repair; (3) applying sophisticated bioinformatic corrections for both FFPE artifacts and batch effects. As comprehensive genomic profiling expands in oncology research, these strategies will enable researchers to leverage the vast archives of clinically annotated FFPE specimens while maintaining the analytical rigor required for robust therapeutic discovery.
Next-generation sequencing (NGS) has revolutionized genomic research, enabling comprehensive detection of genetic variants across the entire genome. However, a significant challenge persists: distinguishing functional variants that contribute to disease mechanisms from passive passenger mutations. Single-omics approaches often provide incomplete biological context, limiting their ability to validate the functional significance of genetic alterations. The integration of transcriptomic and epigenomic data presents a powerful framework to overcome this limitation, creating a synergistic analytical pipeline that connects genetic variation to functional consequences and regulatory mechanisms.
This guide examines how multi-omics integration enhances the accuracy of functional variant validation in chemogenomic screens, comparing experimental methodologies, computational tools, and performance metrics across different approaches. By objectively evaluating how combined transcriptomic-epigenomic analyses outperform single-omics methods, we provide researchers with practical insights for designing robust validation workflows in drug discovery and development.
Foundations of Multi-omics Integration
Defining the Omics Layers
Multi-omics integration combines data from multiple molecular layers to provide a systems-level understanding of biological processes:
- Genomics: Identifies DNA-level alterations including single-nucleotide variants (SNVs), copy-number variations (CNVs), and structural rearrangements [79].
- Epigenomics: Characterizes heritable changes in gene expression not encoded within the DNA sequence itself, including DNA methylation patterns, histone modifications, and chromatin accessibility [79].
- Transcriptomics: Reveals gene expression dynamics through RNA sequencing (RNA-seq), quantifying mRNA isoforms, non-coding RNAs, and fusion transcripts that reflect active transcriptional programs [79].
The Multi-omics Advantage in Functional Validation
Integrating transcriptomic and epigenomic data addresses critical limitations of single-omics approaches for variant validation. While genomic data identifies potential functional variants, it cannot confirm their biological impact. Transcriptomics provides evidence of functional consequences through altered gene expression patterns, while epigenomics reveals the regulatory mechanisms through which these variants operate, such as altered transcription factor binding due to methylation changes [80] [81] [82]. This multi-layered validation is particularly crucial in chemogenomic screens, where understanding how genetic variants influence drug response requires connecting mutations to their functional outcomes and regulatory contexts.
Methodological Approaches and Experimental Designs
Simultaneous Multi-omics Profiling Technologies
SDR-seq (Single-Cell DNA–RNA Sequencing) Recent methodological advances enable simultaneous measurement of multiple omics layers from the same cells, providing inherently matched datasets. SDR-seq simultaneously profiles up to 480 genomic DNA loci and genes in thousands of single cells, enabling accurate determination of coding and noncoding variant zygosity alongside associated gene expression changes [83].
Table 1: SDR-seq Performance Characteristics
| Parameter | Specification | Performance Impact |
|---|---|---|
| Throughput | Thousands of cells | Enables statistical power for rare variant detection |
| Genomic Targets | Up to 480 loci | Flexible panel design for target regions |
| Variant Zygosity | Accurate single-cell determination | Direct linkage of genotype to phenotype |
| Fixation Methods | PFA and glyoxal compared | Glyoxal provides superior RNA target detection |
| Cross-contamination | <0.16% gDNA, 0.8-1.6% RNA | High specificity for both modalities |
The SDR-seq workflow involves several critical steps that ensure data quality. First, cells are dissociated into single-cell suspension, fixed, and permeabilized. In situ reverse transcription is performed using custom poly(dT) primers that add unique molecular identifiers (UMIs), sample barcodes, and capture sequences to cDNA molecules. Cells containing cDNA and gDNA are loaded onto a microfluidics system where droplet generation, cell lysis, and multiplexed PCR amplification of both gDNA and RNA targets occur simultaneously. Distinct overhangs on reverse primers allow separation of NGS library generation for gDNA and RNA, enabling optimized sequencing for each data type [83].
SDR-seq Experimental Workflow
Computational Integration Methods
For datasets generated through separate omics assays, computational integration strategies are required:
Network-Based Approaches Network-based integration methods construct biological networks where nodes represent biomolecules and edges represent functional relationships. By overlaying transcriptomic and epigenomic data onto these networks, researchers can identify regulatory hubs where epigenetic modifications correlate with expression changes of connected genes, prioritizing functional variants that disrupt these relationships [80].
AI-Driven Integration Artificial intelligence approaches, particularly machine learning and deep learning, excel at identifying non-linear patterns across high-dimensional omics spaces. Multi-modal transformers can fuse epigenomic data (e.g., DNA methylation) with transcriptomic data to predict variant impact, while graph neural networks model how variants perturb regulatory networks [79].
Performance Comparison: Multi-omics vs Single-omics Approaches
Validation Accuracy Metrics
Multi-omics integration significantly enhances validation accuracy for functional variants compared to single-omics approaches:
Table 2: Performance Comparison of Variant Validation Methods
| Method | Variant Detection Sensitivity | Functional Interpretation | Regulatory Mechanism Resolution | Application in Complex Disease |
|---|---|---|---|---|
| Genomics Only | High for coding variants | Limited | None | Limited to coding regions |
| Genomics + Transcriptomics | Moderate improvement | Enhanced via expression QTLs | Indirect | Improved for expression-modifying variants |
| Genomics + Epigenomics | High for regulatory variants | Moderate via chromatin states | Direct for regulatory elements | Enhanced for non-coding variants |
| Full Multi-omics | Highest across variant classes | Comprehensive | Direct with functional links | Superior for complex trait dissection |
In a study of ovarian carcinoma, integrated analysis of copy number variation (CNV), DNA methylation, and mRNA expression identified three distinct molecular subtypes with significant survival differences. The iC1 subtype showed lower overall survival and distinct immune cell profiles, which were only detectable through multi-omics integration [81]. This approach identified two genes, UBB and IL18BP, as prognostic biomarkers that would have been missed with single-omics analysis.
Case Study: Neuropsychiatric Disorders
In major depressive disorder (MDD), an integrative analysis combining neuroimaging, brain-wide gene expression from the Allen Human Brain Atlas, and peripheral DNA methylation data revealed that gray matter volume abnormalities were spatially correlated with expression patterns of genes showing differential methylation. This multi-omics approach demonstrated significant associations between decreased gray matter volume and DNA methylation status in the anterior cingulate cortex, inferior frontal cortex, and fusiform face cortex regions [82]. The integrated analysis provided both spatial and biological links between cortical morphological deficits and peripheral epigenetic signatures that would remain undetected in isolated omics analyses.
Experimental Protocols for Multi-omics Validation
Integrated Multi-omics Analysis Protocol
For researchers validating functional variants from chemogenomic screens, the following protocol provides a robust framework:
Step 1: Data Generation and Quality Control
- Perform whole genome or exome sequencing to identify genetic variants
- Conduct RNA sequencing for transcriptomic profiling under relevant conditions
- Execute epigenomic profiling (bisulfite sequencing for methylation, ATAC-seq or ChIP-seq for chromatin accessibility)
- Implement rigorous quality control for each dataset: sequence alignment metrics, coverage uniformity, batch effect detection
Step 2: Data Preprocessing and Normalization
- Normalize expression data using DESeq2 or similar approaches
- Process methylation data using minfi or RnBeads, correcting for cellular heterogeneity
- Annotate genetic variants with functional prediction scores (CADD, SIFT, PolyPhen)
- Perform batch correction using ComBat or similar methods
Step 3: Integrative Analysis
- Identify expression quantitative trait loci (eQTLs) linking variants to expression changes
- Detect methylation quantitative trait loci (meQTLs) connecting variants to methylation changes
- Implement multi-omics factor analysis (MOFA) to identify latent factors across data types
- Construct regulatory networks using WGCNA or similar approaches
Step 4: Functional Validation
- Prioritize variants showing concordant effects across omics layers
- Validate top candidates using CRISPR-based genome editing
- Confirm functional effects through phenotypic assays relevant to chemogenomic context
Multi-omics Variant Validation Pipeline
The Scientist's Toolkit: Essential Research Reagents and Computational Tools
Table 3: Research Reagent Solutions for Multi-omics Studies
| Tool Category | Specific Tools | Function | Application Context |
|---|---|---|---|
| DNA Methylation Analysis | DMRichR, methylKit, RnBeads, ChAMP, MEDIPS | Differential methylation analysis, visualization | BS-seq, RRBS, array-based methylation data |
| Chromatin Analysis | nf-core/chipseq, MACS, BWA, HMCan, deepTools | Peak calling, alignment, quality control | ChIP-seq, ATAC-seq data processing |
| Transcriptomics | nf-core/rnaseq, STAR, Kallisto, DESeq2, EdgeR | Read alignment, quantification, differential expression | RNA-seq, single-cell RNA-seq |
| Multi-omics Integration | FEM, ELMER, Wanderer, Epigenomix, WGCNA | Data integration, regulatory network inference | Combined epigenomic-transcriptomic analysis |
| Variant Calling | DeepVariant, DNAscope, DeepTrio, Clair, Medaka | AI-enhanced variant detection | Germline and somatic variant calling |
| Pathway & Enrichment | GOfuncR, REVIGO, Enrichr, GREAT, STRING | Functional annotation, ontology enrichment | Biological interpretation of findings |
The integration of transcriptomic and epigenomic data represents a paradigm shift in functional variant validation, particularly in the context of chemogenomic screens where understanding the mechanistic basis of drug response variability is crucial. Multi-omics approaches consistently outperform single-omics methods in both validation accuracy and biological insight, enabling researchers to distinguish driver mutations from passenger variants with higher confidence.
As sequencing technologies continue to advance and computational integration methods become more sophisticated, multi-omics will play an increasingly central role in variant interpretation. For research and drug development professionals, adopting these integrated approaches provides a critical advantage in validating therapeutic targets and understanding the complex molecular mechanisms underlying drug responses across diverse genetic backgrounds.
Ensuring Reliability: Benchmarking, Validation, and Performance Metrics
The accurate identification of genetic variants from next-generation sequencing (NGS) data is a foundational requirement in clinical genomics and chemogenomic research. Variant calling pipelines form the analytical core that transforms raw sequencing data into potentially actionable genetic findings, making their accuracy paramount. Inaccurate variant calls can lead to incorrect biological conclusions and flawed clinical interpretations. The establishment of gold standard references has therefore become an essential practice for calibrating these bioinformatics pipelines, ensuring they meet the rigorous demands of clinical and research settings [8] [84].
The Genome in a Bottle (GIAB) Consortium, hosted by the National Institute of Standards and Technology (NIST), has developed into the preeminent resource for benchmark human genomes [84]. By providing comprehensively characterized reference genomes with high-confidence variant calls, GIAB enables objective performance assessment of variant calling pipelines. These benchmark sets are complemented by synthetic datasets like the Synthetic Diploid (Syndip), which are derived from long-read assemblies of homozygous cell lines and provide less biased benchmarking, particularly in challenging genomic regions [8]. Together, these resources provide the foundation for optimizing the accuracy and reliability of NGS analysis in chemogenomic screens and clinical genomics.
Genome in a Bottle (GIAB) Consortium
The GIAB Consortium has developed a technical infrastructure including reference standards, methods, and data to enable clinical translation of whole human genome sequencing. GIAB provides benchmark variant calls for several publicly available human genomes, which are extensively characterized using multiple sequencing technologies and bioinformatics methods [84]. The consortium's priority is the comprehensive characterization of human genomes for benchmarking applications, including analytical validation and technology development.
Key GIAB Reference Samples:
- Pilot Genome: NA12878/HG001 from the HapMap project
- Ashkenazi Jewish Trio: HG002 (son), HG003 (father), HG004 (mother)
- Han Chinese Trio: HG005 (son), HG006 (father), HG007 (mother)
These samples are specifically consented for commercial redistribution, enhancing their utility across both academic and industry settings [84]. For each sample, GIAB provides benchmark variant calls (SNVs and indels) and defines "high-confidence" genomic regions where variant calls can be reliably benchmarked. These resources are continuously updated, with recent expansions including benchmarks for difficult genomic regions, structural variants, and chromosomes X and Y [84].
Synthetic Benchmark Datasets
Synthetic datasets provide an important complementary approach to benchmarking by offering variant calls with known ground truth positions established a priori. The Synthetic Diploid (Syndip) dataset addresses a key limitation of GIAB—the potential for circularity when the same technologies used to create benchmark sets are later evaluated against them [8]. Syndip is derived from de novo long-read assemblies of two homozygous human cell lines, providing particularly valuable benchmarking data for challenging genomic regions such as duplicated sequences [8]. Although the cell lines themselves are not in a public repository, their sequencing datasets are widely available for benchmarking purposes.
Global Alliance for Genomics and Health (GA4GH) Benchmarking Framework
The GA4GH Benchmarking Team, in collaboration with GIAB, has established a best practice framework for variant calling accuracy evaluations [84]. This includes sophisticated comparison tools that account for subtle differences in variant representation, which is particularly important when comparing variant calls against benchmark resources. The framework provides standardized metrics and approaches to ensure consistent and comparable benchmarking across different studies and platforms.
Performance Comparison of Variant Calling Pipelines
Experimental Design for Pipeline Assessment
Rigorous evaluation of variant calling pipelines requires carefully designed experiments that assess performance across different genomic contexts and variant types. A 2022 comprehensive comparison utilized GIAB samples sequenced multiple times to evaluate six different pipeline combinations involving two mapping approaches (GATK with BWA-MEM2 and DRAGEN) and three variant callers (GATK, DRAGEN, and DeepVariant) [34]. Performance was assessed using standard metrics including F1 score (the harmonic mean of precision and recall), precision, and recall, stratified by genomic context and variant type.
Table 1: Comparative Performance of Mapping and Alignment Pipelines
| Mapping Pipeline | Average Runtime (min) | SNV F1 Score | Indel F1 Score | Mendelian Error Rate |
|---|---|---|---|---|
| DRAGEN | 36 ± 2 | 0.9996 | 0.9972 | 0.00047 |
| GATK with BWA-MEM2 | 182 ± 36 | 0.9985 | 0.9883 | 0.00071 |
Table 2: Variant Caller Performance Comparison
| Variant Caller | Runtime (min) | SNV Precision | SNV Recall | Indel Precision | Indel Recall |
|---|---|---|---|---|---|
| DRAGEN | 18 ± 1 | 0.9996 | 0.9996 | 0.9971 | 0.9973 |
| DeepVariant | 231 ± 16 | 0.9998 | 0.9994 | 0.9973 | 0.9956 |
| GATK | 134 ± 20 | 0.9990 | 0.9987 | 0.9902 | 0.9918 |
Performance Across Genomic Contexts
Variant calling performance varies significantly across different genomic contexts, with particular challenges in complex regions. The DRAGEN pipeline demonstrated systematically higher F1 scores, precision, and recall values than GATK with BWA-MEM2 for both SNVs and Indels across all genomic contexts [34]. Differences were most pronounced for recall (sensitivity), representing the probability of detecting true variants.
In difficult-to-map (complex) regions, DRAGEN-based pipelines showed substantial advantages, with F1 scores of 0.9991 for SNVs compared to 0.9964 for GATK-based pipelines. These differences were primarily driven by higher recall values, though precision was also improved [34]. Similar advantages were observed in coding regions, where DRAGEN achieved F1 scores of 0.9996 for SNVs compared to 0.9987 for GATK-based pipelines.
For indels of different sizes, performance differences became more pronounced with increasing variant size. DRAGEN demonstrated better performance than DeepVariant for both insertions and deletions, with advantages growing with indel size [34]. This is particularly relevant for clinical applications where accurate calling of larger indels is often critical for diagnosing genetic disorders.
Diagram 1: Variant Calling Pipeline Benchmarking Workflow. This workflow illustrates the process from raw sequencing data to performance assessment against GIAB truth sets.
Emerging Approaches and Best Practices
Hybrid Sequencing Strategies
Recent advances in sequencing technologies have enabled hybrid approaches that leverage both short-read and long-read sequencing. A 2025 study demonstrated that a hybrid DeepVariant model trained on both Illumina (short-read) and Nanopore (long-read) data can improve germline variant detection accuracy [85]. This approach synergizes the complementary strengths of both technologies: short-read sequencing excels at detecting small variants with high accuracy, while long-read sequencing provides better coverage of complex or repetitive regions.
The hybrid strategy enables shallow hybrid sequencing (combining 15× ONT and 15× Illumina coverage) to achieve accuracy comparable to deeper sequencing with a single technology, potentially reducing overall sequencing costs [85]. This has significant implications for large-scale clinical screening applications where cost-effectiveness is crucial. The approach also enables unified variant calling instead of post hoc merging across platforms, simplifying analytical workflows.
Best Practices for Optimal Performance
Based on comprehensive evaluations of variant calling pipelines, several best practices emerge:
Pre-processing Steps: Read alignment and pre-processing significantly impact variant calling accuracy. Steps including local realignment around indels and base quality score recalibration (BQSR) have been shown to substantially improve call accuracy, with one study demonstrating that realignment and recalibration improved positive predictive value from 35.25% to 88.69% for variants called only after these processing steps [86].
Variant Quality Calibration: The GATK's Variant Quality Score Recalibration (VQSR) generally outperforms hard filtering approaches, building a Gaussian mixture model using annotation values from high-quality variants to evaluate all input variants [86]. While both approaches show high sensitivity (>99.8%), VQSR demonstrates slightly better specificity (99.79% vs. 99.56%) [86].
Joint vs. Individual Variant Calling: For family-based studies, joint variant calling—which processes multiple samples simultaneously—offers advantages over individual calling. Joint calling produces genotypes for all samples at all variant positions, not just positions detected in a given individual, improving the ability to differentiate between true reference calls and insufficient coverage [8].
Diagram 2: Comprehensive Benchmarking Strategy. This diagram shows the multidimensional approach required for thorough pipeline evaluation.
Essential Research Reagents and Computational Tools
Table 3: Key Research Reagent Solutions for Pipeline Calibration
| Resource Type | Specific Examples | Primary Function | Key Features |
|---|---|---|---|
| Reference Samples | GIAB HG001-HG007 | Benchmarking standard | High-confidence variant calls, multiple ancestries |
| Synthetic Datasets | Syndip | Unbiased benchmarking | Known ground truth, especially in difficult regions |
| Alignment Tools | BWA-MEM, DRAGEN, Novoalign | Map reads to reference | Impact downstream variant calling accuracy |
| Variant Callers | GATK, DeepVariant, DRAGEN, Samtools | Identify genetic variants | Differing strengths across variant types |
| Benchmarking Tools | GA4GH Benchmarking Toolsuite | Performance assessment | Standardized comparison against truth sets |
| QC Metrics | Ti/Tv ratio, Mendelian errors | Quality assessment | Indicator of overall variant call quality |
The establishment of comprehensive benchmarking approaches using GIAB and synthetic datasets has transformed the validation of variant calling pipelines for clinical genomics and chemogenomic research. Empirical comparisons demonstrate that pipeline choice significantly impacts variant calling accuracy, with emerging trends favoring DRAGEN for mapping and alignment and both DRAGEN and DeepVariant for variant calling. The development of hybrid strategies that leverage multiple sequencing technologies represents a promising direction for further improving accuracy while potentially reducing costs.
As sequencing technologies continue to evolve and new capabilities like long-read single-molecule sequencing mature, the fundamental importance of rigorous benchmarking using gold standard references remains constant. By implementing the best practices outlined here—utilizing GIAB and synthetic datasets for calibration, evaluating performance across diverse genomic contexts, and employing standardized benchmarking metrics—researchers and clinical laboratories can ensure the highest standards of accuracy for variant detection in both research and clinical applications.
In the field of chemogenomic screens and genomic research, the accuracy of next-generation sequencing (NGS) variant calling is foundational. Key Performance Indicators (KPIs) like sensitivity (recall), precision, and the F-score provide a critical, quantitative framework for evaluating the performance of different variant calling software, directly impacting the reliability of downstream biological conclusions [13] [8]. This guide objectively compares the performance of various variant calling pipelines using published benchmarking data to inform tool selection.
Performance Metrics and Software Comparisons
The table below summarizes the performance of various germline small variant callers from a systematic benchmark of 14 Genome in a Bottle (GIAB) datasets, which provide high-confidence "truth" sets for comparison [17].
Table 1: Performance of Selected Germline Variant Callers on GIAB WES and WGS Data
| Variant Caller | Technology / Basis | Reported SNP F-Score | Reported Indel F-Score | Key Characteristics |
|---|---|---|---|---|
| DeepVariant [17] [87] | AI (Deep Learning) | Consistently high (>0.99) | Consistently high | Highest performance and robustness; uses convolutional neural networks on pileup images. |
| DNAscope [87] | AI (Machine Learning) | High | High | Optimized for speed and accuracy; combines GATK HaplotypeCaller with machine learning genotyping. |
| Clair3 [17] [87] | AI (Deep Learning) | High | High | Fast performance, particularly strong at lower sequencing coverages. |
| Strelka2 [17] | Statistical | High | High | Strong performance, though more dependent on input data quality than DeepVariant. |
| GATK HaplotypeCaller [17] [8] | Statistical (Haplotype) | High | High | Well-established, widely used benchmark in germline variant calling. |
| FreeBayes [17] | Statistical (Haplotype) | Lower than top performers | Lower than top performers | Often shows lower accuracy compared to more modern tools. |
For commercial software that does not require programming expertise, a 2025 benchmark on GIAB exome data revealed the following performance metrics [18]:
Table 2: Performance of Commercial, No-Code Variant Calling Software
| Software | Variant Calling Engine | Precision (SNV/Indel) | Recall (SNV/Indel) | Runtime (for 3 samples) |
|---|---|---|---|---|
| Illumina DRAGEN | DRAGEN Enrichment | >99% / >96% | >99% / >96% | 29 - 36 minutes |
| CLC Genomics | Lightspeed Germline | High | High | 6 - 25 minutes |
| Partek Flow | GATK HaplotypeCaller | High | High | 3.6 - 29.7 hours |
| Partek Flow | FreeBayes & Samtools (Union) | Lower (especially for indels) | Lower (especially for indels) | 3.6 - 29.7 hours |
Structural variant (SV) callers exhibit significantly different performance profiles, as they identify larger genomic alterations. A 2024 benchmark study evaluated caller performance on whole-genome sequencing data from GIAB samples [88].
Table 3: Performance of Structural Variant (SV) Callers
| SV Caller | Deletion F-Score | Duplication F-Score | Insertion F-Score | Inversion F-Score |
|---|---|---|---|---|
| Manta | ~0.50 (Highest) | <0.20 | ~0.20 (Highest) | Low |
| Delly | ~0.35 | <0.20 | ~0.05 | Low |
| GridSS | ~0.25 | <0.20 | ~0.05 | Low |
| Sniffles | ~0.15 | <0.20 | ~0.05 | Low |
Experimental Protocols for Benchmarking
A standardized and rigorous experimental protocol is essential for generating comparable and trustworthy benchmarking data.
The Benchmarking Workflow
The following diagram illustrates the standard workflow for benchmarking variant callers, from data input to KPI calculation.
Key Methodological Components
- Gold Standard Datasets: The Genome in a Bottle (GIAB) Consortium develops reference materials and high-confidence variant calls for several human genomes (e.g., HG001, HG002) [18] [17] [8]. These "truth sets" are derived from the integration of multiple sequencing technologies and bioinformatics methods, providing a reliable standard against which to benchmark.
- High-Confidence Regions: Benchmarking is performed within well-characterized "high-confidence" genomic regions defined by GIAB. This ensures that discrepancies are true errors and not ambiguities in the truth set itself [17] [89].
- Standardized Comparison Tools: The GA4GH Benchmarking Tool (hap.py) is a community-standard software for comparing variant calls. It uses sophisticated comparison algorithms (e.g.,
vcfeval) to handle complex variant representations, ensuring accurate matching and counting of True Positives (TPs), False Positives (FPs), and False Negatives (FNs) [18] [89]. - KPI Calculation: The tool outputs counts that are used to calculate the primary KPIs:
- Precision = TP / (TP + FP) measures the accuracy of positive predictions.
- Recall (Sensitivity) = TP / (TP + FN) measures the ability to find all true variants.
- F-Score = 2 * (Precision * Recall) / (Precision + Recall) provides a single metric balancing precision and recall [89].
The Scientist's Toolkit
The table below details key reagents, software, and data resources essential for conducting a variant calling benchmark or analysis.
Table 4: Essential Research Reagents and Resources for Variant Calling
| Item Name | Type | Critical Function in Experiment |
|---|---|---|
| GIAB Reference DNA & Data [18] [8] | Biological Reference Material | Provides the physical sample and associated sequencing data with known, high-confidence variants to serve as the "ground truth" for benchmarking. |
| Agilent SureSelect [18] | Exome Capture Kit | Used in the preparation of the sequencing libraries for the GIAB WES data, defining the genomic regions interrogated. |
| BWA-MEM Aligner [18] [17] [8] | Bioinformatics Software | The widely adopted standard tool for aligning NGS short reads to a reference genome (e.g., GRCh38). |
| GA4GH Benchmarking Tools (hap.py) [18] [89] | Bioinformatics Software | The standardized software for comparing a query VCF against a truth set to calculate precision, recall, and F-score. |
| Variant Call Format (VCF) | Data Format | The standard, structured text file format used to store gene sequence variations. Serves as the input and output for all comparison tools. |
Strategic Implications for Tool Selection
Choosing an appropriate variant caller requires balancing KPIs with other practical considerations.
- AI-Powered vs. Traditional Callers: AI-based tools like DeepVariant and Clair3 consistently achieve top-tier accuracy in benchmarks for small variants [17] [87]. However, they can be computationally intensive. Traditional, highly optimized statistical callers like GATK or Strelka2 remain excellent, high-performance choices [17] [8].
- Commercial "No-Code" Solutions: Platforms like Illumina DRAGEN and CLC Genomics Workbench offer exceptional accuracy and speed with user-friendly interfaces, making powerful analysis accessible to wet-lab scientists and smaller clinics without dedicated bioinformatics support [18].
- The "Best Tool" Depends on Variant Type: No single caller is optimal for all variant types. As shown in Table 3, Manta excels at detecting deletions but performs poorly on other SVs. For a comprehensive view, consolidating variant calls from multiple specialized tools is often necessary [13] [88].
- Impact of Sequencing Depth: KPI values are not static and can be influenced by sequencing depth. For SV callers, performance generally improves with higher depth, but beyond a point (e.g., >100x), precision may decrease due to a surge in false positive calls, underscoring the need for depth-appropriate tool selection [88].
For researchers and drug development professionals, the accuracy of Next-Generation Sequencing (NGS) variant calling in chemogenomic screens is paramount. This technical accuracy, however, operates within a critical framework of regulatory compliance and quality management. The Clinical Laboratory Improvement Amendments (CLIA) establish the foundational quality standards for U.S. clinical laboratories, ensuring testing reliability and patient safety [90]. Recent 2025 CLIA updates have introduced significant changes, including tightened personnel qualifications and enhanced proficiency testing (PT) criteria, which directly impact how laboratories implement and validate NGS workflows [90] [91]. Simultaneously, initiatives like the CDC/APHL Next-Generation Sequencing Quality Initiative (NGS QI) develop specific tools to help laboratories build robust Quality Management Systems (QMS) that satisfy these evolving regulatory requirements while managing the inherent complexities of NGS technology [92]. This guide explores this intersection, providing a comparative analysis of NGS variant calling performance data within the context of modern QMS and CLIA compliance.
Regulatory Framework: CLIA & Quality Management Systems
Key 2025 CLIA Updates and Their Impact on NGS Labs
The 2025 CLIA updates represent the first major overhaul in decades, raising the bar for laboratory compliance [90]. For laboratories utilizing NGS, these changes have several critical implications:
- Digital-Only Communication: CMS has phased out paper mailings in favor of exclusive electronic communication. Labs must ensure contact details are accurate and monitored to avoid missing critical notices, which is essential for maintaining accreditation and staying informed on policy changes affecting NGS workflows [90].
- Updated Personnel Qualifications: The rules tighten requirements for lab directors and staff. Certain degrees and "board eligibility only" no longer automatically qualify, meaning labs may need to update job descriptions and documentation for bioinformaticians and other specialized NGS roles [90] [91]. While existing staff may be grandfathered in, reviewing personnel files is crucial for compliance.
- Stricter Proficiency Testing (PT) Criteria: Standards for PT are stricter, with some newly regulated analytes added. Labs must review their PT programs to ensure their quality systems align with updated expectations, directly affecting the ongoing verification of NGS test accuracy [90].
- Announced Audits: Accrediting bodies like the CAP can now announce inspections up to 14 days in advance. This necessitates that laboratories, including those running complex NGS pipelines, be inspection-ready at all times, with comprehensive and current documentation [90].
The NGS Quality Initiative (NGS QI) as a QMS Framework
In response to the challenges of implementing NGS in clinical and public health settings, the CDC and APHL formed the NGS QI. Its mission is to address common challenges associated with personnel, equipment, and process management by providing publicly available tools and resources for building a robust QMS [92]. A core principle of the NGS QI is that a proper QMS enables continual improvement and proper document management. The initiative develops tools that are crosswalked with regulatory, accreditation, and professional bodies (e.g., FDA, CMS, CAP) to ensure they provide current and compliant guidance [92]. Key resources include the QMS Assessment Tool, Identifying and Monitoring NGS Key Performance Indicators SOP, NGS Method Validation Plan, and the NGS Method Validation SOP [92]. These documents assist laboratories in navigating the complex validation process governed by CLIA, which has increased in complexity due to sample type variability, intricate library preparation, and evolving bioinformatics tools [92].
Structured Guidance for NGS Test Lifecycle
Complementing the NGS QI resources, the College of American Pathologists (CAP), in collaboration with other professional organizations, has created a set of structured worksheets that guide the user through the entire life cycle of an NGS test [93]. These seven worksheets provide a practical, step-by-step approach to QMS implementation for NGS, covering phases from initial design to routine operation and quality management [93].
Diagram 1: The relationship between regulatory frameworks, QMS tools, and NGS data quality.
Comparative Analysis of NGS Variant Calling Pipelines
The selection of a bioinformatics pipeline is a critical decision within a laboratory's QMS, directly impacting the accuracy and reliability of variant calling in chemogenomic research. The following section provides a structured comparison of popular pipelines, highlighting their performance in key metrics.
Performance Benchmarking of Variant Calling Pipelines
A comprehensive 2022 empirical study compared six whole genome sequencing pre-processing pipelines, involving two mapping and alignment approaches and three variant callers, using Genome in a Bottle (GIAB) reference samples [34]. The study assessed performance using F1 score (the harmonic mean of precision and recall), precision, and recall for both SNVs and Indels.
Table 1: Performance Benchmarking of Variant Calling Pipelines (based on HG002 sample)
| Mapping & Alignment | Variant Caller | F1 Score (SNVs) | F1 Score (Indels) | Precision (SNVs) | Recall (SNVs) | Runtime (mins) |
|---|---|---|---|---|---|---|
| DRAGEN | DRAGEN | High | Highest | High | High | 36 ± 2 |
| DRAGEN | DeepVariant | Highest | High | Highest | Medium | 256 ± 7 |
| GATK BWA-MEM2 | GATK | Low | Low | Low | Low | ≥ 180 |
| DRAGEN | GATK | High | High | High | High | ~189 |
Data adapted from Scientific Reports volume 12, Article number: 21502 (2022) [34]
The study concluded that mapping and alignment play a key role in variant calling, with the DRAGEN pipeline systematically outperforming GATK with BWA-MEM2. It showed higher F1 scores, precision, and recall for both SNVs and Indels [34]. In the variant calling step, DRAGEN and DeepVariant performed similarly and both were superior to GATK, with slight advantages for DRAGEN for Indels and for DeepVariant for SNVs [34]. DRAGEN was also the fastest pipeline by a significant margin.
Performance in Genomic Regions of Varying Complexity
The performance of variant callers is not uniform across the genome. The same study stratified performance based on region complexity, a critical consideration for chemogenomic screens that may target diverse genomic contexts.
Table 2: Performance in Difficult-to-Map (Complex) vs. Simple-to-Map Regions
| Pipeline | F1 Score SNVs (Simple) | F1 Score SNVs (Complex) | F1 Score Indels (Simple) | F1 Score Indels (Complex) |
|---|---|---|---|---|
| DRAGEN-DRAGEN | High | High | Highest | Highest |
| DRAGEN-DeepVariant | High | High | High | High |
| GATK-GATK | Low | Low | Low | Low |
Data adapted from Scientific Reports volume 12, Article number: 21502 (2022) [34]
For SNVs, F1 scores were substantially lower in complex regions when GATK was used for mapping and alignment compared to any DRAGEN-based pipeline. These differences were primarily caused by low recall values for GATK-based pipelines, meaning they failed to detect a larger number of true variants in these challenging regions [34]. This demonstrates the superiority of modern pipelines in maintaining accuracy across the entire genome.
The Rise of Deep Learning and Long-Read Sequencing
The benchmarking evidence for the superiority of deep learning-based variant callers extends beyond human genomics to bacterial systems. A 2024 study in eLife evaluated variant calling on Oxford Nanopore Technologies (ONT) sequence data across 14 bacterial species [20]. The findings revealed that deep learning-based variant callers, particularly Clair3 and DeepVariant, significantly outperformed traditional methods and even exceeded the accuracy of Illumina sequencing, especially when applied to ONT’s super-high accuracy model [20]. ONT's superior performance was attributed to its ability to overcome Illumina’s errors, which often arise from difficulties in aligning reads in repetitive and variant-dense genomic regions [20]. This is particularly relevant for chemogenomic screens in microbial systems or for human genomes in regions of high complexity or homology.
Furthermore, cross-platform benchmarking of SARS-CoV-2 sequencing revealed that while Illumina NovaSeq produced the highest depth of coverage and completeness, implementing proper quality controls on long-read data from ONT MinION and PacBio Sequel II achieved consistent lineage assignments across all platforms [50]. This underscores the importance of robust bioinformatic quality control within the QMS.
Diagram 2: A generalized NGS bioinformatics workflow showing key stages and QMS oversight points.
Experimental Protocols for Benchmarking and Validation
For a laboratory to validate an NGS pipeline under CLIA regulations, a rigorous experimental design is required. The following protocol is synthesized from the methodologies of the cited benchmarking studies.
Protocol for Cross-Pipeline Variant Calling Benchmarking
This protocol is designed to objectively compare the accuracy of different bioinformatic pipelines, providing the experimental data needed for initial validation under a QMS.
1. Sample Selection and Truth Set Definition:
- Reference Materials: Use well-characterized reference samples with established "truth sets." The Genome in a Bottle (GIAB) consortium provides human genomic DNA reference materials (e.g., HG002) with high-confidence variant calls [34]. For bacterial genomics, create a pseudo-real truthset by applying real variants from a donor genome to a sample's reference assembly [20].
- Sequencing: Sequence the selected sample(s) multiple times to account for run-to-run variability. The 2022 WGS study sequenced one GIAB sample 70 times in different runs [34]. Use the same DNA extraction for all sequencing platforms to avoid bias.
2. Data Processing with Target Pipelines:
- Pipelines: Select pipelines for comparison. A typical design includes:
- Mapping/Alignment: GATK (with BWA-MEM2) vs. DRAGEN.
- Variant Calling: GATK HaplotypeCaller vs. DRAGEN vs. DeepVariant.
- Execution: Process the raw FASTQ files from the sequencing runs through each pipeline combination (e.g., DRAGEN mapping + DRAGEN calling, DRAGEN mapping + DeepVariant calling, etc.). Ensure all pipelines use the same reference genome.
3. Performance Metric Calculation:
- Variant Comparison: Use tools like
hap.pyorvcfevalto compare the pipeline-generated VCF files against the known truth set. - Key Metrics: Calculate for each pipeline and for different variant types (SNVs, Indels):
- Precision: (True Positives) / (True Positives + False Positives). Measures the fraction of reported variants that are real.
- Recall (Sensitivity): (True Positives) / (True Positives + False Negatives). Measures the fraction of real variants that are detected.
- F1 Score: 2 * (Precision * Recall) / (Precision + Recall). The harmonic mean of precision and recall.
- Stratified Analysis: Calculate these metrics in different genomic contexts: simple-vs-complex regions, coding-vs-non-coding regions, and for Indels of different sizes [34].
4. Additional Assessments:
- Mendelian Consistency: For trio data, calculate the Mendelian inheritance error fraction to assess consistency in familial inheritance patterns [34].
- Computational Efficiency: Record the total run time and computational resources (CPU, memory) required for each pipeline.
Protocol for Long-Read Variant Caller Benchmarking in Bacteria
For labs using long-read technologies for bacterial chemogenomics, the following adapted protocol is relevant.
1. Sample and Truth Set Preparation:
- Bacterial Strains: Select a diverse panel of Gram-positive and Gram-negative bacterial species with varying GC content [20].
- Truth Set Generation: For each sample, select a closely related donor genome (~99.5% Average Nucleotide Identity). Identify all variants between the sample and donor using tools like
minimap2andMUMmer. Apply this high-confidence variant set to the sample's reference to create a mutated reference genome, which serves as the truth set [20].
2. Sequencing and Basecalling:
- Platforms: Sequence the same DNA extraction using both Illumina (for a gold-standard comparison) and ONT.
- ONT Models: Basecall the ONT data using different accuracy models (e.g., fast, high-accuracy (hac), and super-accuracy (sup)) and read types (simplex and duplex) [20].
3. Variant Calling and Evaluation:
- Caller Selection: Evaluate a mix of deep learning-based (e.g., Clair3, DeepVariant) and traditional variant callers.
- Performance Analysis: Compare the variants called from each ONT dataset (and the Illumina dataset) against the established truth set. Calculate precision and recall to determine the most accurate combination of basecalling model and variant caller [20].
The Scientist's Toolkit: Essential Research Reagent Solutions
The implementation and validation of NGS workflows require a combination of physical reagents, data resources, and software tools. The following table details key components essential for ensuring accuracy and compliance.
Table 3: Essential Reagents, Resources, and Tools for NGS Variant Calling
| Category | Item | Function and Application |
|---|---|---|
| Reference Materials | Genome in a Bottle (GIAB) Reference Samples | Provides a gold-standard set of genomic DNA with highly validated variant calls for benchmarking and validating NGS pipeline accuracy [34]. |
| Bioinformatics Software | DRAGEN Pipeline | An integrated, highly efficient bioinformatics platform for mapping, alignment, and variant calling, noted for its high speed and accuracy [34]. |
| GATK with BWA-MEM2 | A traditional, widely used pipeline for mapping and variant calling, often used as a baseline for performance comparisons [34]. | |
| DeepVariant | A deep learning-based variant caller that converts the variant calling problem into an image classification task, known for high precision [34] [20]. | |
| Quality Management | CAP NGS Worksheets | A set of seven structured worksheets guiding the entire lifecycle of a clinical NGS test, from design and validation to routine operation and quality management [93]. |
| NGS QI Validation Plan/SOP | Tools from the CDC/APHL NGS Quality Initiative providing templates and guidance for performing NGS method validation in a CLIA-compliant manner [92]. | |
| Data Resources | GIAB Truth Sets | Community-developed sets of high-confidence variant calls for GIAB reference genomes, used as a benchmark for evaluating variant caller performance [34]. |
| CLSI MM09 Guideline | A professional guideline providing recommendations for designing, validating, and managing clinical tests based on NGS and Sanger sequencing [93]. |
The accurate identification of somatic variants is a cornerstone of precision oncology, enabling the matching of patients with targeted therapies based on the genetic profile of their tumor. Next-generation sequencing (NGS) has made comprehensive genomic profiling feasible in clinical and research settings. However, the rapidly evolving landscape of sequencing platforms and bioinformatics pipelines introduces a critical variable: the choice of analytical tools can significantly impact variant identification, potentially altering clinical interpretations and patient outcomes [94] [95]. This case study systematically compares the performance of various sequencing technologies and variant calling pipelines, evaluating their concordance and accuracy in detecting actionable cancer mutations within the context of chemogenomic screen research. We provide researchers and drug development professionals with a data-driven guide to inform pipeline selection, thereby enhancing the reliability of genomic data used to identify novel therapeutic targets and biomarkers.
Comparative Analysis of Sequencing Platforms & Bioinformatics Pipelines
Sequencing Platform Performance
Sequencing platforms from both Illumina and BGI were evaluated for use in Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS). Performance was consistent across major platforms, with all showing high base quality suitable for robust variant detection [94].
Table 1: Sequencing Platform Performance Metrics for Germline Variant Calling [94]
| Sequencing Platform | Application | Average Read Depth | Q20 (%) | Q30 (%) |
|---|---|---|---|---|
| BGISEQ500 | WES | 328.4X | >95 | >89 |
| MGISEQ2000 | WES | 129.4X | >95 | >89 |
| HiSeq4000 | WES | 395.17X | >95 | >89 |
| NovaSeq | WES | 241.52X | >95 | >89 |
| BGISEQ500 | WGS | 41.03X | >92 | >83 |
| MGISEQ2000 | WGS | 45.13X | >92 | >83 |
| HiSeq4000 | WGS | 58.0X | >92 | >83 |
| NovaSeq | WGS | 28.96X | >92 | >83 |
| HiSeq Xten | WGS | 38.93X | >92 | >83 |
A separate 2024 study focusing on Targeted Sequencing (TS) using the TruSight Oncology 500 (TSO500) panel corroborates these findings, reporting that platforms including NovaSeq 6000, NextSeq 550, MGISEQ-2000, GenoLab M, and FASTASeq 300 all produced high-quality data, with Q20 scores exceeding 94% and deep, uniform coverage (>2000x) [95].
Variant Calling Pipeline Performance
The accuracy of variant calling pipelines was benchmarked against the GIAB consortium's gold-standard dataset for sample NA12878 [94]. Performance was measured using the F-score, the harmonic mean of precision and recall.
Table 2: Performance (F-score) of Variant Calling Pipelines Across Platforms [94]
| Sequencing Platform | Application | Variant Type | GATK-HC | Strelka2 | Samtools-Varscan2 |
|---|---|---|---|---|---|
| BGISEQ500 | WES | SNP | 0.98 | 0.99 | 0.96 |
| MGISEQ2000 | WES | SNP | 0.98 | 0.99 | 0.96 |
| HiSeq4000 | WES | SNP | 0.98 | 0.99 | 0.96 |
| NovaSeq | WES | SNP | 0.98 | 0.99 | 0.96 |
| BGISEQ500 | WES | INDEL | 0.83 | 0.91 | 0.75 |
| MGISEQ2000 | WES | INDEL | 0.82 | 0.90 | 0.76 |
| HiSeq4000 | WES | INDEL | 0.83 | 0.91 | 0.75 |
| NovaSeq | WES | INDEL | 0.83 | 0.91 | 0.76 |
For targeted sequencing in oncology, a 2024 benchmarking study evaluated five bioinformatics pipelines (HaplotypeCaller, Mutect2, SNVer, VarScan 2, and SiNVICT) on data from multiple platforms. The study found that the FASTASeq 300 platform analyzed with SNVer and VarScan 2 algorithms achieved the highest sensitivity (100%) and precision (100%) for calling high-confidence variants in the OncoSpan reference standard [95]. Furthermore, SNVer and VarScan 2 consistently performed best for both SNP and InDel sensitivity across different sample types, including cell-free DNA (cfDNA) and cancer cell lines [95].
The Rise of AI-Based Variant Callers
A new generation of variant callers leveraging artificial intelligence (AI) has demonstrated transformative potential. These tools use deep learning models to analyze sequencing data, often achieving superior accuracy compared to traditional methods [16].
Table 3: Overview of AI-Based Variant Calling Tools [16]
| Tool | Underlying Technology | Key Features | Strengths | Limitations |
|---|---|---|---|---|
| DeepVariant | Deep Convolutional Neural Network (CNN) | Analyzes pileup images of aligned reads; supports short and long reads. | High accuracy, eliminates need for manual filtering; used in UK Biobank. | High computational cost. |
| DeepTrio | Deep CNN | Extends DeepVariant for family trio analysis (child and parents). | Improved accuracy in challenging regions and de novo mutation detection. | High computational cost. |
| DNAscope | Machine Learning (ML) | Combines GATK HaplotypeCaller with an AI-based genotyping model. | High speed and accuracy, reduced computational cost vs. DeepVariant/GATK. | Not a deep learning model; requires no GPU. |
| Clair/Clair3 | Deep CNN | Specializes in both short-read and long-read data. | High performance at lower coverages; fast runtime. | Earlier versions inaccurate for multi-allelic variants. |
These AI-based tools have shown strong performance in benchmarking studies. However, their computational demands and the "black box" nature of their decisions are important considerations for clinical and research implementation [16].
Experimental Protocols for Systematic Benchmarking
To ensure reproducible and reliable results, benchmarking studies follow rigorous experimental protocols. The following workflow details the standard methodology for comparing variant calling pipelines.
Key Research Reagent Solutions
The following reagents and materials are essential for conducting a robust pipeline comparison study.
Table 4: Essential Research Reagents and Materials for Benchmarking
| Item | Function / Description | Example Use Case |
|---|---|---|
| Reference Standard DNA | Well-characterized, cell-line-derived DNA with a known truth set of variants. Essential for benchmarking. | GIAB NA12878 for germline variants; OncoSpan HD832 (contains 386 variants in 152 cancer genes) for somatic variants [94] [95]. |
| Targeted Sequencing Panel | A probe set used to enrich for specific genomic regions of interest, such as cancer-related genes. | TruSight Oncology 500 (TSO500) panel covers 523 cancer-related genes for detecting SNPs, InDels, CNVs, and fusions [95]. |
| Library Prep Kit | Reagents for fragmenting DNA, attaching adapters, and amplifying libraries for sequencing. | TSO500 library preparation kit or similar (e.g., TargetSeq One for cfDNA) [95]. |
| Bioinformatics Pipelines | A suite of software tools for sequence alignment, variant calling, and filtering. | GATK, Strelka2, VarScan 2, SNVer, and AI-based callers like DeepVariant and DNAscope [94] [95] [16]. |
Impact on Identification of Actionable Cancer Mutations
Clinical Context of Actionable Mutations
In clinical oncology, the goal of genomic profiling is to identify "actionable" mutations—genetic alterations for which an approved or investigational targeted therapy exists [96]. A study of 500 patients with advanced cancer found that 30% harbored such potentially actionable alterations, with prevalence varying significantly by tumor type [96]. For example, while pancreatic cancers had a high rate of KRAS mutations, other actionable targets like BRAF in melanoma and PIK3CA in breast cancer were also common [96]. This underscores the importance of sensitive and comprehensive detection methods.
Pipeline Choice Influences Mutation Detection
The choice of variant calling pipeline directly impacts the sensitivity and specificity of mutation detection, which can subsequently influence the identification of patients eligible for targeted therapies. The benchmarking studies reveal that while overall concordance is often high, the divergence in variant identification, particularly for InDels, can be substantial [94]. For instance, in WES data, the F-score for InDel calling ranged from 0.75 to 0.91 depending on the pipeline, meaning that a suboptimal pipeline could miss a significant number of true InDels or introduce false positives [94].
Furthermore, a multi-platform, multi-pipeline study on targeted sequencing recommended integrating calls from multiple tools (e.g., SNVer and VarScan 2) to improve overall sensitivity and accuracy for the cancer genome [95]. This integrative approach helps mitigate the limitations of any single pipeline, ensuring that potentially critical, low-frequency mutations are not overlooked.
A Decision Framework for Pipeline Selection
The following diagram synthesizes the findings of this case study into a logical framework for selecting an optimal variant calling strategy based on research or clinical objectives.
This case study demonstrates that the selection of sequencing platforms and bioinformatics pipelines is not a mere technical detail but a critical factor that directly influences the accuracy and completeness of actionable mutation detection in cancer genomics. Systematic comparisons reveal that while modern platforms from Illumina and BGI generate high-quality data, the choice of variant caller—from established tools like Strelka2 to emerging AI-powered solutions like DeepVariant and DNAscope—significantly impacts results, especially for challenging variants like InDels. For clinical chemogenomic screens and research aimed at drug discovery, adopting a rigorously benchmarked, and potentially integrated, pipeline is essential. This ensures the reliable genomic data needed to identify genuine therapeutic targets, correlate genotypes with drug response, and ultimately advance the field of precision oncology.
Conclusion
Accurate NGS variant calling is the critical linchpin connecting chemogenomic screens to actionable biological insights and viable therapeutic targets. A successful strategy requires a holistic approach that integrates foundational best practices in data pre-processing, careful selection and combination of bioinformatic tools, and relentless optimization guided by rigorous benchmarking. The future points toward increasingly automated, AI-driven pipelines that seamlessly incorporate multi-omics data and long-read sequencing, offering a more comprehensive view of the genomic landscape. By adopting these principles, researchers can significantly enhance the precision and reproducibility of their findings, ultimately accelerating the pace of drug discovery and the advancement of precision medicine.
References
- 1. Comparative evaluation of SNVs, indels, and structural ... [https://www.nature.com/articles/s41439-024-00276-x]
- 2. Validation of genetic variants from NGS data using deep ... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-023-05255-7]
- 3. Benchmarking reveals superiority of deep learning variant ... [https://elifesciences.org/reviewed-preprints/98300]
- 4. Comparisons of performances of structural variants detection ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0314982]
- 5. CSN and CAVA: variant annotation tools for rapid, robust next ... [https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-015-0195-6]
- 6. Genome-wide functional annotation of variants: a systematic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11911558/]
- 7. Methods to improve the accuracy of next-generation sequencing [https://pmc.ncbi.nlm.nih.gov/articles/PMC9895957/]
- 8. Best practices for variant calling in clinical sequencing [https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-020-00791-w]
- 9. A Bioinformatics Toolkit for Next-Generation Sequencing in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10741970/]
- 10. What are the best pipelines for NGS (Next Generation ... [https://synapse.patsnap.com/article/what-are-the-best-pipelines-for-ngs-next-generation-sequencing-data-analysis]
- 11. Next-Generation Sequencing Technology: Current Trends ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10376292/]
- 12. Next-Generation Sequencing (NGS) in Drug Discovery & ... [https://www.corning.com/worldwide/en/products/life-sciences/resources/stories/the-cutting-edge/integrating-next-generation-sequencing-into-drug-discovery.html]
- 13. Summoning insights: NGS variant calling best practice [https://www.ogt.com/ca/resources/ngs-resources-and-support/ngs-resources/summoning-insights-ngs-variant-calling-best-practices/]
- 14. Best practices for variant calling in clinical sequencing - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7586657/]
- 15. StratoMod: predicting sequencing and variant calling errors ... [https://www.nature.com/articles/s42003-024-06981-1]
- 16. Artificial intelligence in variant calling: a review - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12055765/]
- 17. Systematic benchmark of state-of-the-art variant calling ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-022-08365-3]
- 18. Benchmarking of variant calling software for whole-exome ... [https://www.nature.com/articles/s41598-025-97047-7]
- 19. A Novel and Accelerated Method for Integrated Alignment ... [https://www.frontiersin.org/journals/bioinformatics/articles/10.3389/fbinf.2025.1691056/full]
- 20. Benchmarking reveals superiority of deep learning variant ... [https://elifesciences.org/articles/98300]
- 21. Discovery and characterization of artifactual mutations in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3616734/]
- 22. Characterization and mitigation of artifacts derived from NGS ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-024-10157-w]
- 23. Benchmarking UMI-aware and standard variant callers for low ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-024-10737-w]
- 24. Transformation of alignment files improves performance of ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-02923-y]
- 25. Next-Generation Sequencing Methodologies To Detect ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10843083/]
- 26. Correcting errors in PCR-derived libraries for rare allele ... [https://www.nature.com/articles/s42003-025-08537-3]
- 27. Establishing a Variant Allele Frequency Cutoff for Manual ... [https://www.sciencedirect.com/science/article/pii/S1525157824002502]
- 28. A Novel Variant Calling Assay for Low Frequency Variants [https://www.mdpi.com/2073-4425/12/4/507]
- 29. Performance optimization in DNA short-read alignment [https://pmc.ncbi.nlm.nih.gov/articles/PMC10060706/]
- 30. Aligner optimization increases accuracy and decreases ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5643015/]
- 31. BWA MEM vs BOWTIE2 , which is best? [https://www.biostars.org/p/293397/]
- 32. Comparison of seven SNP calling pipelines for the next- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8803190/]
- 33. Fast and accurate DNASeq variant calling workflow composed ... [https://humgenomics.biomedcentral.com/articles/10.1186/s40246-024-00666-w]
- 34. Comparison of calling pipelines for whole genome ... [https://www.nature.com/articles/s41598-022-26181-3]
- 35. Benchmarking accelerated next-generation sequencing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12092081/]
- 36. Next-generation sequencing of newborn screening genes [https://www.nature.com/articles/s41525-020-00142-z]
- 37. Leveraging long-read sequencing technologies for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12075302/]
- 38. Fast, accurate, and racially unbiased pan-cancer tumor ... [https://www.nature.com/articles/s41698-022-00340-1]
- 39. Germline contamination and leakage in whole genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5793408/]
- 40. Clinical application of tumour-in-normal contamination ... [https://www.nature.com/articles/s41467-023-44158-2]
- 41. A benchmarking study of individual somatic variant callers ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11790059/]
- 42. A Simplified Approach to Somatic Analysis in NGS [https://www.euformatics.com/blog-post/a-simplified-approach-to-somatic-analysis-in-ngs]
- 43. Population-based metrics to stratify somatic variant calling ... [https://www.illumina.com/science/genomics-research/articles/population-metrics-stratify-somatic-variant.html]
- 44. Determination of high-confidence germline genetic variants in ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-11889-z]
- 45. Read trimming has minimal effect on bacterial SNP-calling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8116680/]
- 46. Negligible effects of read trimming on the accuracy... [https://f1000research.com/articles/13-506]
- 47. Comparison of Read Mapping and Variant Calling Tools for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7238416/]
- 48. Accuracy and efficiency of germline variant calling ... [https://www.nature.com/articles/s41598-020-77218-4]
- 49. Short-Read Sequencing vs. Long ... [https://seqwell.com/short-read-sequencing-vs-long-read-sequencing-which-technology-is-right-for-your-research/]
- 50. Short-Read and Long-Read Whole Genome Sequencing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12031342/]
- 51. comparison of large-scale chemogenomic fitness signatures [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-022-08395-x]
- 52. Long-read sequencing vs short-read sequencing [https://frontlinegenomics.com/long-read-sequencing-vs-short-read-sequencing/]
- 53. DNA Sequencing: How to Choose the Right Technology [https://frontlinegenomics.com/dna-sequencing-how-to-choose-the-right-technology/]
- 54. Methodological Comparison of Short-Read and Long ... [https://www.mdpi.com/1422-0067/26/18/9254]
- 55. A Hitchhiker's Guide to long-read genomic analysis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12047252/]
- 56. Optimal sequencing depth for measuring the concentrations of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12390757/]
- 57. Advances on Recognizing and Managing Tumor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6136997/]
- 58. 肿瘤异质性与个体化治疗未来展望篇 [http://www.caca.org.cn/system/2023/10/11/030048152.shtml]
- 59. PrecisionOncology WhitePaper 0725 CN-web - Life Science [https://collateral.meridianlifescience.com/view/216014212]
- 60. 次世代定序技術引領精準醫療新紀元 [https://geneonline.news/%E6%AC%A1%E4%B8%96%E4%BB%A3%E5%AE%9A%E5%BA%8F%E6%8A%80%E8%A1%93%E5%BC%95%E9%A0%98%E7%B2%BE%E6%BA%96%E9%86%AB%E7%99%82%E6%96%B0%E7%B4%80%E5%85%83/]
- 61. NGS变局下,基因测序的匠心之选——HiFi测序! [https://www.ebiotrade.com/newsf/2025-2/202522195737479.htm]
- 62. [J]. 中国癌症防治杂志, 2024, 16(6): 638-649. [http://www.caca.org.cn/system/2025/01/09/030124432.shtml]
- 63. Performance analysis of conventional and AI-based variant ... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-023-05596-3]
- 64. Comparison of GATK and DeepVariant by trio sequencing [https://www.nature.com/articles/s41598-022-05833-4]
- 65. DeepChem-Variant: A Modular Open Source Framework ... [https://arxiv.org/html/2411.11513v2]
- 66. DeepVariant Blog - Google [https://google.github.io/deepvariant/]
- 67. DeepSomatic AI: 98% accuracy in cancer mutation detection [https://www.rdworldonline.com/googles-deepsomatic-ai-achieves-98-accuracy-in-cancer-mutation-detection-outperforms-standard-tools/]
- 68. Genome-scale chemogenomic CRISPR screens in human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7868615/]
- 69. Quality control, modeling, and visualization of CRISPR ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0843-6]
- 70. A critical spotlight on the paradigms of FFPE-DNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10415133/]
- 71. Fresh frozen vs FFPE samples for next-generation ... [https://www.lexogen.com/blog/fresh-frozen-vs-ffpe-samples-for-next-generation-sequencing-studies/]
- 72. Comparison of Clinical Targeted Next-Generation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4912568/]
- 73. Large-scale analysis of whole genome sequencing data ... [https://www.nature.com/articles/s41467-024-51577-2]
- 74. Alternative Tissue Fixation Protocols Dramatically Reduce ... [https://www.sciencedirect.com/science/article/pii/S0023683723002234]
- 75. Guidelines for Validation of Next-Generation Sequencing– ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6941185/]
- 76. Identifying and mitigating batch effects in whole genome ... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1756-z]
- 77. Comparative analysis of batch correction methods for FDG ... [https://www.nature.com/articles/s41598-023-45296-9]
- 78. Batch Effect Correction [https://www.10xgenomics.com/analysis-guides/introduction-batch-effect-correction]
- 79. AI-driven multi-omics integration in precision oncology [https://pmc.ncbi.nlm.nih.gov/articles/PMC12634751/]
- 80. Integrating Multi-Omics Data: Methods and Applications in ... [https://www.sciencedirect.com/science/article/pii/S2215017X25000657]
- 81. Integrated multi-omics analysis of genomics, epigenomics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6629004/]
- 82. Integrative omics analysis reveals epigenomic and ... [https://www.nature.com/articles/s41398-023-02724-8]
- 83. Functional phenotyping of genomic variants using joint ... [https://www.nature.com/articles/s41592-025-02805-0]
- 84. Genome in a Bottle | NIST [https://www.nist.gov/programs-projects/genome-bottle]
- 85. Joint processing of long- and short-read sequencing data ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12296420/]
- 86. Validation and assessment of variant calling pipelines for next ... [https://humgenomics.biomedcentral.com/articles/10.1186/1479-7364-8-14]
- 87. Artificial intelligence in variant calling: a review [https://www.frontiersin.org/journals/bioinformatics/articles/10.3389/fbinf.2025.1574359/full]
- 88. Comparison of structural variant callers for massive whole ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-024-10239-9]
- 89. Best Practices for Benchmarking Germline Small Variant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6699627/]
- 90. What 2025 CLIA Updates Mean for Labs and Compliance [https://reesscientific.com/blog/2025-clia-updates-for-labs]
- 91. 2025 Changes to Point-of-Care Testing Regulatory ... [https://myadlm.org/education/point-of-care-testing-professional-certification/the-point/articles/2025/may]
- 92. The Next-Generation Sequencing Quality Initiative and ... [https://wwwnc.cdc.gov/eid/article/31/13/24-1175_article]
- 93. Next Generation Sequencing (NGS) Worksheets [https://www.cap.org/member-resources/precision-medicine/next-generation-sequencing-ngs-worksheets]
- 94. Systematic comparison of germline variant calling ... [https://www.nature.com/articles/s41598-019-45835-3]
- 95. Systematic comparison of variant calling pipelines of target ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10794554/]
- 96. Clinical next generation sequencing to identify actionable ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4652990/]