Breaking the Barrier: Outer Membrane Permeability as the Key to Overcoming Gram-Negative Antibiotic Resistance
This article provides a comprehensive analysis of the Gram-negative bacterial outer membrane as a formidable permeability barrier and a major contributor to antibiotic resistance.
Breaking the Barrier: Outer Membrane Permeability as the Key to Overcoming Gram-Negative Antibiotic Resistance
Abstract
This article provides a comprehensive analysis of the Gram-negative bacterial outer membrane as a formidable permeability barrier and a major contributor to antibiotic resistance. Aimed at researchers and drug development professionals, it explores the foundational biology of the asymmetric outer membrane, detailing the roles of lipopolysaccharides (LPS) and porins. It further examines methodological approaches for enhancing compound penetration, troubleshooting common resistance phenotypes, and validating strategies through comparative studies across pathogens like Pseudomonas aeruginosa and Acinetobacter baumannii. By synthesizing foundational knowledge with recent advances in permeabilization and efflux inhibition, this review aims to inform the rational design of next-generation antimicrobials effective against multidrug-resistant Gram-negative pathogens.
The Gram-Negative Fortress: Deconstructing the Outer Membrane Barrier
The lipid bilayer constitutes the fundamental barrier of the cell, but its performance is not merely a function of its bulk properties. Compositional asymmetry—the non-random distribution of lipid species between the two leaflets of the bilayer—is a highly conserved and energetically costly feature of the plasma membrane in eukaryotes and the outer membrane in Gram-negative bacteria. This review posits that asymmetry is a critical structural adaptation that optimizes the membrane for the conflicting demands of forming a robust permeability barrier while enabling efficient cellular signaling and, in bacteria, conferring intrinsic antibiotic resistance. We synthesize recent advances demonstrating that the exoplasmic or outer leaflet is specialized for low permeability, rich in saturated lipids and cholesterol (or lipopolysaccharides in bacteria), forming a tightly packed, ordered phase. In contrast, the cytoplasmic or inner leaflet is enriched in unsaturated lipids, creating a more fluid, disordered environment that facilitates the diffusion and interaction of signaling molecules. This guide provides a quantitative dissection of this model, detailing the experimental and computational methodologies driving this field forward, and frames the implications for overcoming membrane-based antibiotic resistance in Gram-negative pathogens.
Biological membranes are not symmetric. The plasma membrane of eukaryotic cells and the outer membrane of Gram-negative bacteria exhibit profound lipid asymmetry, a nonequilibrium state where the lipid composition of the outer (exoplasmic) and inner (cytoplasmic) leaflets are distinct [1] [2]. Cells invest substantial free energy, in the form of tens to hundreds of ATP hydrolysis events per translocated lipid, to establish and maintain this asymmetry via active transporters like flippases and floppases [1]. Such a significant energetic investment implies a critical functional benefit.
The core hypothesis of this article is that lipid asymmetry represents an evolutionary solution to a fundamental design challenge: the membrane must be both an impermeable barrier and a fluid matrix for dynamic processes. This guide will explore the structural basis of how asymmetry resolves this conflict, with a specific focus on the outer membrane of Gram-negative bacteria as a primary determinant of antibiotic resistance. The asymmetric outer membrane, with its lipopolysaccharide (LPS)-rich outer leaflet, provides a "formidable barrier" that restricts the passive influx of antibiotics, making these pathogens particularly challenging to treat [3] [4] [5].
Structural and Compositional Foundations of the Asymmetric Barrier
The two leaflets of the cellular plasma membrane have distinct lipid compositions tailored for their specific functions. This asymmetry is a hallmark of plasma membranes across eukaryotes and many prokaryotes [1].
The Exoplasmic (Outer) Leaflet: This leaflet is predominantly enriched in lipids with saturated fatty acyl chains, such as sphingomyelin and phosphocholine lipids. These lipids pack tightly due to their straight hydrocarbon chains, forming a dense, liquid-ordered phase. In Gram-negative bacteria, this role is taken to an extreme with the exclusive presence of lipopolysaccharide (LPS) in the outer leaflet of the outer membrane. The complex, bulky structure of LPS, anchored by lipid A, creates a nearly impenetrable hydrophobic barrier [3] [4] [5].
The Cytoplasmic (Inner) Leaflet: This leaflet is primarily composed of lipids with (poly-)unsaturated fatty acids, such as the aminophospholipids phosphatidylserine (PS) and phosphatidylethanolamine (PE). The kinks in the unsaturated hydrocarbon chains prevent tight packing, resulting in a liquid-disordered phase that is more fluid and dynamic [1] [6].
Table 1: Characteristic Lipid Compositions of Asymmetric Membrane Leaflets
| Leaflet | Representative Lipid Components | Physical State | Primary Functional Assignment |
|---|---|---|---|
| Exoplasmic/Outer | Sphingomyelin, Phosphocholine, Lipopolysaccharide (LPS in bacteria) | Liquid-ordered phase | Impermeable Barrier Function |
| Cytoplasmic/Inner | Phosphatidylserine (PS), Phosphatidylethanolamine (PE), Polyunsaturated lipids | Liquid-disordered phase | Signaling & Molecular Dynamics |
This compositional divide is not merely a static separation; it creates a composite material with emergent properties. The two leaflets can be viewed as acting as two resistances in series, where the overall permeability is dominated by the least permeable leaflet [1]. This conceptual framework is illustrated in Figure 1.
Figure 1: The two leaflets of the asymmetric plasma membrane form a composite material, with the outer leaflet providing a tight barrier and the inner leaflet enabling fluidity for signaling.
Quantitative Basis of Permeability
The barrier function of a membrane is quantitatively described by its permeability coefficient (P), governed by the Meyer-Overton rule: P = K·D/d, where K is the partition coefficient, D is the diffusion constant within the membrane, and d is the membrane thickness [1]. For an asymmetric bilayer, the total permeability (P~total~) is the reciprocal sum of the permeabilities of the individual leaflets, acting as resistances in series: 1/P~total~ = 1/P~o.l.~ + 1/P~i.l.~ (where o.l. and i.l. denote outer and inner leaflets) [1].
When the permeability difference between leaflets is large, the overall membrane permeability is effectively determined by the less permeable leaflet: P~total~ ≈ P~o.l.~. This is precisely the case in the cellular plasma membrane, where the tightly packed outer leaflet dominates barrier function.
Experimental data robustly supports this model. A comparative study using liposomes with lipid compositions mimicking the exoplasmic versus cytoplasmic leaflets found that the cytoplasmic-mimic membranes were 18 to 90 times more permeable to various polar substances [1]. This dramatic difference is attributed to key physical properties:
- Area Per Lipid: This is a dominant factor controlling permeability, particularly for water. The saturated lipids and high cholesterol content of the exoplasmic leaflet minimize the area per lipid, creating a densely packed structure that is difficult for solutes to penetrate [1].
- Membrane Phase: The exoplasmic leaflet's ability to form a liquid-ordered phase is critical. Water permeability in a fluid-phase DPPC bilayer can be 2000 times lower than in a gel-phase bilayer, and a 7-fold difference persists between liquid-ordered and liquid-disordered phases [1].
Table 2: Quantitative Permeability Differences Between Leaflet Compositions
| Parameter | Exoplasmic/Outer Leaflet Mimic | Cytoplasmic/Inner Leaflet Mimic | Experimental Basis |
|---|---|---|---|
| Relative Permeability | 1X (Baseline) | 18X to 90X higher | Liposome studies with polar substances [1] |
| Phase State | Liquid-ordered (L~o~) | Liquid-disordered (L~d~) | Fluorescent packing reporters [1] |
| Key Physical Trait | Small area per lipid, high packing density | Large area per lipid, low packing density | Molecular dynamics & biophysical studies [1] [6] |
| Cholesterol Content | High | ~1/2 to 1/3 of L~o~ phase | Domain formation studies [1] |
In Gram-negative bacteria, the barrier is even more extreme. The outer membrane's permeability to hydrophilic antibiotics is largely governed by porin channels, as the LPS layer itself is a formidable barrier to hydrophobic molecules [3] [5]. Modifications to LPS structure, such as the addition of 4-amino-4-deoxy-L-arabinose to lipid A, can further reduce its negative charge and decrease the binding and uptake of cationic antimicrobial peptides and antibiotics like polymyxin, constituting a major resistance mechanism [5].
Methodologies for Investigating Asymmetric Membranes
Studying asymmetric bilayers is methodologically challenging, as traditional model membranes are often symmetric. Below are key experimental and computational approaches driving progress in the field.
Computational Modeling with Molecular Dynamics (MD)
MD simulations provide atomic-resolution insights into membrane properties and are an indispensable tool for studying asymmetry [6]. A critical challenge is the initial construction of the asymmetric bilayer model, as the chosen protocol heavily influences the results, particularly regarding differential stress—a non-zero leaflet tension arising from mismatched leaflet properties [6].
Table 3: Protocols for Constructing Asymmetric Bilayers in MD Simulations
| Construction Protocol | Core Principle | Best Suited For |
|---|---|---|
| Equal Numbers (EqN) | Ensure an equal number of lipid molecules in each leaflet. | Initial studies, systems with minimal compositional disparity. |
| Surface Area (SA) | Match the leaflet surface areas to those from cognate symmetric bilayers. | Investigating properties sensitive to lateral pressure and packing. |
| Zero Differential Stress (0-DS) | Adjust lipid numbers to achieve zero leaflet tension (τ = 0). | Simulating a relaxed, natural membrane state; studying mechanical properties. |
| P21 Boundary Conditions | Use specialized periodic boundaries that allow lipid flip-flop between leaflets. | Studying systems where slow lipid exchange is relevant. |
The choice of construction method is paramount, as it determines the presence and magnitude of differential stress, which in turn can affect membrane properties like thickness, area per lipid, and the lateral diffusion of lipids and proteins [6]. The general workflow for this computational analysis is summarized in Figure 2.
Figure 2: Workflow for molecular dynamics simulation of asymmetric lipid bilayers, highlighting the critical step of selecting a bilayer construction protocol.
Experimental Assessment of Barrier Permeability
In both biological and model membrane systems, quantifying permeability is essential. In clinical and preclinical settings, Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE-MRI) is a gold standard for assessing blood-brain barrier (BBB) permeability, a key example of a specialized barrier function [7].
Advanced DCE-MRI methods involve acquiring T1-weighted images before and after intravenous injection of a Gadolinium-based contrast agent (Gd-DTPA). The "post-pre comparison" method involves a pixel-wise statistical comparison (e.g., t-test with false discovery rate correction) between pre- and post-contrast scans. Pixels with statistically significant intensity changes within a defined enhancement range (calibrated using reference tissues like muscle and eyeball) are identified as regions with a leaky BBB [7]. This method provides a semi-quantitative assessment of barrier integrity with a less demanding imaging protocol.
The Scientist's Toolkit: Key Reagents and Methods
| Research Tool / Reagent | Function in Research | Key Context |
|---|---|---|
| Asymmetric Liposomes/Vesicles | Model membranes with controlled, asymmetric lipid distribution in each leaflet. | Essential for in vitro study of true membrane asymmetry, enabling permeability and diffusion assays [2]. |
| Molecular Dynamics (MD) Software (e.g., GROMACS, CHARMM, NAMD) | Simulates atomic-level dynamics of asymmetric bilayers over time. | Allows investigation of lipid-lipid and lipid-protein interactions, differential stress, and permeability pathways [6]. |
| Fluorescent Lipid Analogs & Lipid-Anchored Proteins | Probes for measuring lateral diffusion and dynamics in the membrane leaflets. | Used in FRAP and other live-cell imaging techniques to demonstrate higher fluidity in the cytoplasmic leaflet [1]. |
| General Diffusion Porins (e.g., OmpF, OmpC) | Bacterial outer membrane proteins forming water-filled channels for hydrophilic solute influx. | Critical for studying antibiotic uptake in Gram-negative bacteria; mutations here are a common resistance mechanism [3] [4]. |
| Lipopolysaccharide (LPS) | Primary component of the outer leaflet of the Gram-negative bacterial outer membrane. | Key reagent for modeling the formidable permeability barrier of bacteria and studying resistance mechanisms like LPS modification [3] [5]. |
| Gadolinium-Based Contrast Agents (e.g., Gd-DTPA) | Tracer for in vivo permeability assessment using imaging techniques like DCE-MRI. | Used to quantitatively evaluate the integrity of biological barriers like the Blood-Brain Barrier (BBB) [7]. |
The asymmetric bilayer is not a passive, static wall but a sophisticated, actively maintained composite material. Its structural design elegantly solves the fundamental problem of integrating a resilient, impermeable barrier with a dynamic, fluid signaling platform. In the context of antibiotic resistance, the Gram-negative outer membrane represents a perfected example of this principle, where extreme asymmetry and unique molecular components like LPS create a formidable defense.
Future research will continue to deepen our quantitative understanding of interleaflet coupling—how the physical state of one leaflet influences the other—and its role in cellular physiology and drug resistance. Advancing experimental methods to create more complex and tunable asymmetric model membranes, combined with increasingly powerful molecular dynamics simulations, will be crucial [2]. Furthermore, a detailed molecular understanding of the outer membrane permeability barrier opens the door to rational drug design strategies aimed at bypassing or disrupting this barrier, thereby resensitizing resistant pathogens to conventional antibiotics. The study of membrane asymmetry thus stands as a critical frontier at the intersection of cell biology, biophysics, and pharmaceutical science.
Lipopolysaccharide (LPS) is the defining molecular component of the outer membrane (OM) of most Gram-negative bacteria [8]. This complex glycolipid fulfills a critical dual function: it provides crucial structural integrity to the bacterial cell while simultaneously forming a formidable permeability barrier that protects against external threats, including many antimicrobial compounds [9] [8]. The effectiveness of this barrier, particularly its ability to exclude hydrophobic molecules, is a direct consequence of the unique chemical structure of LPS and its precise packing within the membrane [8] [10]. Understanding the relationship between LPS composition, membrane packing, and the resulting hydrophobic exclusion is fundamental to research aimed at overcoming innate antibiotic resistance in Gram-negative pathogens.
Structural Composition of LPS
An LPS molecule is architecturally divided into three distinct domains, each contributing specific properties to the overall function of the OM.
- Lipid A: This is the hydrophobic anchor of the LPS molecule, embedding it in the outer leaflet of the OM [11] [9]. Lipid A is a conserved glucosamine-based disaccharide that is phosphorylated and typically acylated with four to seven saturated fatty acids [9] [8] [10]. This domain is the endotoxic principle of LPS, primarily responsible for triggering a potent immune response in hosts through the TLR4/MD-2 receptor complex [11] [12].
- Core Oligosaccharide: Attached to lipid A, this domain consists of a short chain of sugars, including unusual ones like keto-deoxyoctulosonate (Kdo) and heptose [11] [9]. The core region can be subdivided into the inner and outer core. The inner core is often phosphorylated or substituted with charged molecules like phosphoethanolamine, contributing significantly to the overall negative charge of the OM [9] [10].
- O-Antigen (O-Ag): This is a highly variable polymer of repeating oligosaccharide units that extends distally from the core [11]. The presence and length of the O-antigen determine whether LPS is classified as "smooth" (with O-Ag) or "rough" (without O-Ag or with a truncated core) [11] [8]. The O-antigen is a key virulence factor, protecting bacteria from host immune responses such as complement-mediated lysis [8].
Table 1: Domains of the LPS Molecule and Their Key Characteristics
| Domain | Chemical Nature | Function | Variability |
|---|---|---|---|
| Lipid A | Hydrophobic; glucosamine disaccharide with saturated acyl chains | Membrane anchor; endotoxic activity; permeability barrier | Conserved, but modifications (acylation, phosphorylation) occur and impact immune activation [9] [8]. |
| Core Oligosaccharide | Hydrophilic; contains Kdo, heptoses, hexoses | Structural integrity; contributes to membrane charge | Moderately variable; truncations create "rough" phenotypes [11] [8]. |
| O-Antigen | Highly hydrophilic; repeating sugar units | Protects from host defenses; serological specificity; adhesion | Highly variable; defines serotypes; length is polymorphic [11] [8]. |
LPS Packing and the Hydrophobic Barrier
The exceptional ability of the OM to exclude hydrophobic toxins and antibiotics is not due to a typical phospholipid bilayer but arises from the unique physicochemical properties of LPS and its tight packing in the outer leaflet [8] [10].
Molecular Basis of the Barrier
The barrier function is a direct result of two key characteristics of the LPS layer:
- Low Membrane Fluidity: The lipid A domain typically contains saturated fatty acids (often 6 in E. coli), which engage in extensive lateral interactions with neighboring LPS molecules. This results in a membrane with very low fluidity, creating a tightly packed, quasi-crystalline surface that is difficult for hydrophobic molecules to penetrate [8] [10].
- Dense Packing via Electrostatic Cross-linking: The LPS molecule carries multiple negative charges from phosphate and carboxyl groups on lipid A and the core oligosaccharide [8]. Strong electrostatic repulsion between these charges would normally prevent dense packing. This repulsion is neutralized by divalent cations (e.g., Mg²⁺, Ca²⁺) that intercalate between adjacent LPS molecules, forming ionic bridges that dramatically enhance the packing density and stability of the OM [8] [10].
Impact of LPS Structure on Packing and Permeability
The precise structure of LPS directly influences the integrity of the permeability barrier.
- Core Oligosaccharide Length and Charge: Mutants with truncated LPS cores ("deep rough" mutants) exhibit a markedly more permeable OM [10]. A shorter core reduces the distance over which electrostatic repulsion acts, weakening the cross-bridging by divalent cations [13]. Furthermore, a reduction in core length and charge has been experimentally shown to increase bacterial sensitivity to hydrophobic solvents like butanol, as it compromises OM integrity [13].
- O-Antigen Length: Contrary to traditional understanding, recent research indicates that the presence of a long O-antigen polysaccharide can compromise the OM barrier to antibiotics. Transport and assembly of the bulky O-antigen at the cell surface appears to create defects that increase permeability, revealing a critical trade-off for bacteria between protection from host immunity and maintaining membrane integrity [14].
The following diagram illustrates how LPS structure and composition contribute to its barrier function.
Diagram 1: The relationship between LPS structure, its molecular properties, and the resulting hydrophobic barrier function. The O-antigen's role is complex, as its assembly can sometimes disrupt the dense packing.
Role in Hydrophobic Exclusion and Antibiotic Resistance
The OM asymmetric bilayer, with its dense LPS matrix, is exceptionally effective at hindering the penetration of hydrophobic antibiotics, which are often potent against Gram-positive bacteria [10]. This intrinsic resistance mechanism is termed hydrophobic exclusion.
- Mechanism of Exclusion: In a conventional phospholipid bilayer, hydrophobic molecules can dissolve and diffuse through the hydrophobic core. The tightly packed, hydrophilic surface of the LPS layer prevents this. The saturated acyl chains of lipid A create a rigid, ordered environment, while the extensive sugar network of the core and O-antigen presents a polar shell, making the initial entry of hydrophobic compounds thermodynamically unfavorable [8] [10].
- Evidence from Mutants: The critical role of a full-length, charged LPS core is demonstrated by "deep rough" mutants. These strains, which produce severely truncated LPS, show a dramatic increase in sensitivity to hydrophobic antibiotics (e.g., novobiocin, fusidic acid), macrolides, and dyes because their OM is compromised and contains patches of phospholipids that are more easily traversed [10].
- Bacterial Countermeasures to Host Defenses: Bacteria can further refine their LPS barrier to resist host-derived antimicrobial peptides (CAMPs). A common modification is the addition of cationic groups like 4-amino-4-deoxy-L-arabinose (Ara4N) or phosphoethanolamine to the phosphate groups on lipid A [12] [10]. This reduces the net negative charge of the OM, decreasing the initial electrostatic attraction of cationic peptides and resulting in a more tightly packed, impermeable LPS layer that is resistant to compounds like polymyxin B [10].
Table 2: LPS Modifications and Their Impact on Antibiotic Permeability
| LPS Modification | Mechanism | Effect on OM Permeability | Resistance Conferred |
|---|---|---|---|
| Truncation of Core (Rough mutants) | Reduced charge and length; incorporation of phospholipids in outer leaflet [10]. | Greatly Increased | Increased sensitivity to hydrophobic antibiotics (e.g., novobiocin, fusidic acid, macrolides) [10]. |
| Ara4N/PEtn addition to Lipid A | Neutralizes negative charge on phosphates; enhances packing [12] [10]. | Decreased | Resistance to cationic antimicrobial peptides (e.g., Polymyxin B) [10]. |
| Alteration of O-Antigen Length | Long O-Ag may disrupt efficient packing and transport during assembly [14]. | Context-dependent increase | Shorter O-Ag can improve resistance to some antibiotics by improving barrier integrity [14]. |
| Reduction of Acyl Chains (e.g., to penta/tetra-acylated) | Alters geometry and packing density of Lipid A [8] [15]. | Can decrease immune recognition (evasion) but may alter fluidity [8]. | Evasion of TLR4-mediated immune response; variable effect on antibiotic resistance [8] [15]. |
Experimental Methods for Analysis
Research on LPS-driven hydrophobic exclusion relies on a suite of biochemical, genetic, and biophysical techniques.
Genetic Manipulation of LPS Structure
A core methodology involves the creation and analysis of bacterial mutants with defined alterations in their LPS.
- Objective: To directly test the contribution of specific LPS domains (O-antigen, core, lipid A) to OM barrier function.
- Protocol:
- Strain Construction: Use targeted gene knockouts (e.g., of waa genes involved in core biosynthesis or wba genes for O-antigen synthesis) to generate isogenic strains that produce truncated "rough" LPS or lack the O-antigen entirely [14] [10].
- Phenotypic Analysis:
- Antibiotic Sensitivity Assay: Perform minimum inhibitory concentration (MIC) assays using a range of hydrophobic (e.g., novobiocin, erythromycin) and hydrophilic antibiotics. Rough mutants are expected to show significantly lower MICs for hydrophobic drugs [10].
- Chemical Sensitivity Assay: Test sensitivity to detergents (e.g., SDS) and bile salts, which also indicate OM integrity [10].
- LPS Characterization: Verify the LPS structure of mutants using SDS-PAGE and silver staining, which reveals the size and ladder-like pattern of smooth LPS versus the truncated bands of rough LPS [14].
Molecular Dynamics (MD) Simulations
In silico modeling provides atomic-level insights into how LPS packing creates a barrier.
- Objective: To simulate the dynamic behavior of the asymmetric OM and quantify the energetics of solute penetration.
- Protocol:
- Membrane Model Construction: Build a computational model of the OM bilayer with LPS in the outer leaflet and phospholipids in the inner leaflet. Models can include LPS with varying core lengths and charges [13].
- Simulation Setup: Place the model in a solvated box with ions (including Mg²⁺) and energy-minimize it. Run production simulations under controlled temperature and pressure.
- Analysis:
- *Lateral Packing and Fluidity: Calculate the lateral spacing between LPS molecules and the order parameter of the lipid A acyl chains. Tighter packing and higher order indicate a better barrier [13].
- Free Energy Calculations: Use umbrella sampling or similar methods to calculate the potential of mean force (PMF) for the translocation of a hydrophobic probe (e.g., butanol) across the OM. A high energy barrier indicates effective hydrophobic exclusion [13].
Permeabilization Assays
These assays use agents that specifically disrupt LPS packing to sensitize bacteria to antibiotics.
- Objective: To experimentally compromise the LPS layer and measure the resultant increase in antibiotic susceptibility.
- Protocol:
- Treatment with Chelators: Use EDTA to chelate the divalent cations (Mg²⁺, Ca²⁺) that cross-bridge LPS molecules. This disrupts electrostatic stabilization, increases membrane permeability, and leads to the release of some LPS [10].
- Treatment with Cationic Peptides: Use polymyxin B nonapeptide (PMBN), a derivative that lacks direct bactericidal activity but retains the ability to bind LPS and displace cross-bridging cations. This "permeabilizes" the OM without killing the cell [10].
- Checkerboard Assay: Combine sub-lethal concentrations of a permeabilizer (EDTA or PMBN) with a dilution series of an antibiotic. A synergistic reduction in the MIC of the antibiotic confirms the role of the OM as a permeability barrier [10].
The workflow for a comprehensive experimental analysis of LPS-mediated barrier function is summarized below.
Diagram 2: A combined experimental and computational workflow for analyzing the role of LPS in hydrophobic exclusion.
The Scientist's Toolkit: Key Research Reagents
The following table lists essential reagents and tools used in LPS and OM permeability research.
Table 3: Key Reagents for LPS and Outer Membrane Research
| Reagent / Tool | Function / Application | Key Characteristics |
|---|---|---|
| Polymyxin B Nonapeptide (PMBN) | OM permeabilizer; used to sensitize cells to hydrophobic antibiotics by disrupting LPS packing [10]. | Lacks the fatty acid tail of polymyxin B, thus non-bactericidal but retains LPS-binding activity [10]. |
| EDTA (Ethylenediaminetetraacetic acid) | Metal chelator; strips Mg²⁺/Ca²⁺ from LPS, disrupting ionic cross-links and increasing permeability [10]. | Commonly used in combination assays with antibiotics or detergents to test OM integrity. |
| Deep Rough Mutant Strains (e.g., E. coli K-12 derivatives, Salmonella Re mutants) | Models for studying the effect of severely truncated LPS cores on OM permeability and antibiotic resistance [14] [10]. | Produce lipooligosaccharide (LOS) lacking O-antigen and most of the core; highly sensitive to hydrophobic drugs [10]. |
| Anti-LPS Antibodies | Detection and serotyping of LPS; used in Western blot, ELISA, and immunofluorescence. | Specific to the O-antigen polysaccharide; allows for identification of bacterial serovars [8]. |
| TLR4/MD-2 Reporter Cell Lines (e.g., HEK-Blue hTLR4) | In vitro assessment of the endotoxic activity (immunostimulatory potential) of different LPS or lipid A structures [16] [15]. | Cells engineered to express the human TLR4/MD-2 complex and an inducible reporter gene for NF-κB activation. |
| LpxC Inhibitors (e.g., CHIR-090) | Potent, specific inhibitors of the LpxC enzyme; used to study the essentiality of LPS and as a starting point for novel antibiotic development [12]. | Targets the first committed step in lipid A biosynthesis; bactericidal for most Gram-negative bacteria [12]. |
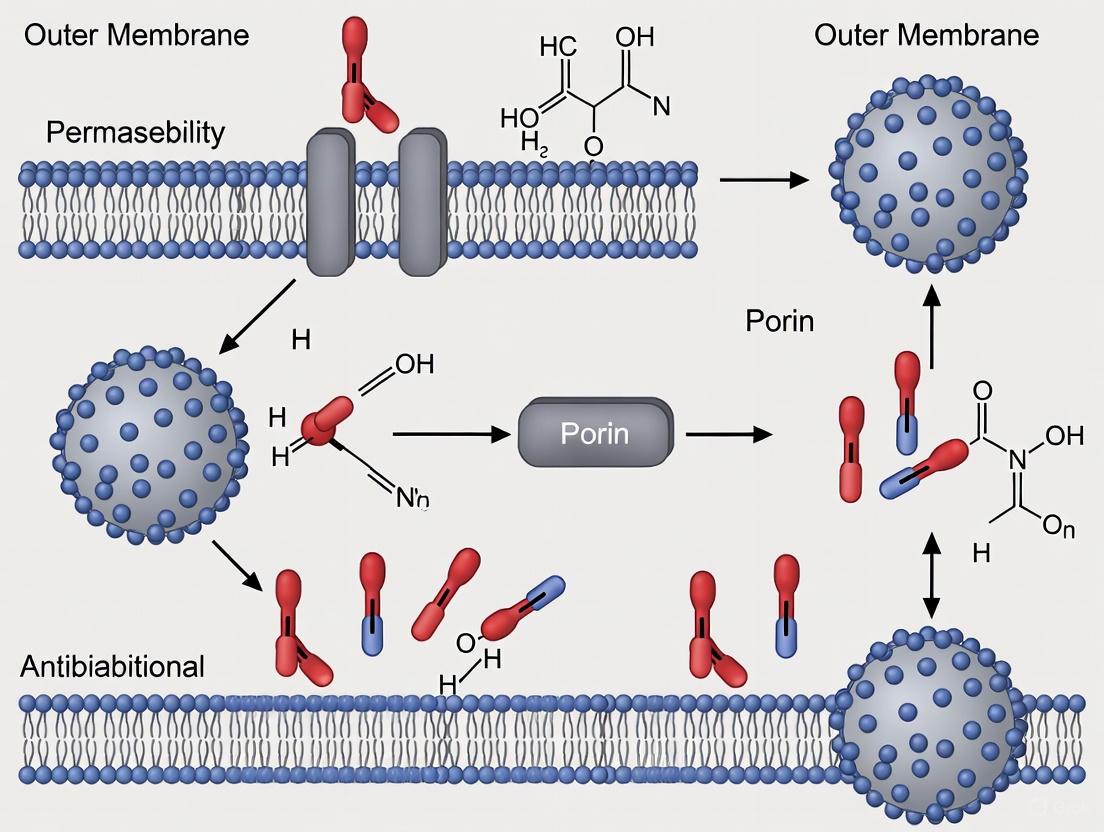
The outer membrane (OM) of Gram-negative bacteria constitutes a formidable permeability barrier, serving as a primary intrinsic resistance mechanism against antimicrobial agents [17] [18]. This asymmetric lipid bilayer, with its lipopolysaccharide (LPS)-enriched outer leaflet, provides exceptional protection while still allowing the selective passage of essential nutrients and ions [19]. The gatekeepers of this selective permeability are porins—integral membrane proteins that form water-filled channels facilitating the passive diffusion of hydrophilic molecules across the otherwise impermeable OM [19] [20]. Porins fundamentally function as molecular sieves, with pore diameters typically restricting passage to molecules under 600 Da, thereby balancing the membrane's protective role with the cellular need for nutrient uptake and waste expulsion [21].
The clinical significance of porins extends far beyond their basic physiological functions, as they represent critical determinants in bacterial pathogenesis and antibiotic resistance [19]. Modifications in porin expression, structure, or function can dramatically alter bacterial susceptibility to multiple antibiotic classes, positioning these proteins at the forefront of antimicrobial resistance research [20] [17]. The escalating global health threat posed by multidrug-resistant Gram-negative pathogens has intensified scientific interest in understanding porin biology, with the ultimate goal of developing novel therapeutic strategies that either exploit or counteract porin-mediated permeability mechanisms [18].
This technical guide comprehensively examines the structural and functional dichotomy between general diffusion porins and substrate-specific channels, detailing their respective roles in hydrophilic influx. By integrating recent advances in porin research with established foundational knowledge, we provide researchers with a sophisticated framework for understanding how porin pathways influence antibiotic permeability and resistance phenotypes, ultimately informing future drug development efforts against resistant Gram-negative infections.
Structural Architecture of Porin Channels
The β-Barrel Fold: A Conserved Structural Motif
Porins exhibit a characteristic transmembrane β-barrel architecture that distinguishes them from the α-helical bundles typical of inner membrane proteins [19] [21]. This structural motif consists of 14 to 22 antiparallel β-strands arranged in a cylindrical formation, with the first and last strands connected in an antiparallel fashion to complete the barrel closure [19]. The β-strands themselves are amphipathic, featuring an alternating pattern of hydrophobic residues facing the lipid bilayer and hydrophilic residues lining the central aqueous pore [22] [21]. This strategic arrangement creates a thermodynamically stable interface with the membrane environment while facilitating the passage of water-soluble compounds through the channel interior.
The dimensions of the porin β-barrel are remarkably conserved, with a height of approximately 25-30 Å corresponding to the thickness of the lipid bilayer, and an oval cross-section with diameters ranging from 30-35 Å laterally [19]. A key structural feature stabilizing porins within the membrane is the presence of aromatic girdles—clusters of tyrosine and phenylalanine residues—positioned at both the extracellular and periplasmic membrane interfaces [19]. These girdles serve as molecular anchors, with tyrosine predominating at the extracellular face and phenylalanine at the periplasmic side, creating distinct aromatic symmetry that enhances structural integrity [19].
The Constriction Zone: Molecular Basis for Selectivity
Perhaps the most functionally significant structural element within porins is the constriction zone, which dictates the size exclusion limit and charge selectivity of the channel [19] [17]. This narrow region is formed primarily by the inward folding of the third extracellular loop (L3), which bends back into the barrel at approximately half the height of the channel [19]. The constriction zone creates the narrowest part of the pore, effectively determining the molecular weight cutoff for permeating solutes—typically 600 Da or less for general porins in Enterobacterales [18].
The molecular discrimination at the constriction zone arises from both steric and electrostatic factors. The physical dimensions of this region create a molecular sieve that excludes molecules based on size, while the specific arrangement of charged amino acid residues along the constriction lining establishes an electrostatic field that preferentially attracts or repels solutes based on their charge [17]. For instance, in the general porin OmpF, the constriction zone features clustered acidic residues that create a net negative charge, weakly favoring cation permeation [21]. In contrast, specific porins like PhoE contain positively charged residues that enhance anion selectivity [22].
Table 1: Structural Classification of Major Porins in Gram-Negative Bacteria
| Porin Type | β-Strand Count | Oligomeric State | Exclusion Limit (Da) | Representative Examples | Key Structural Features |
|---|---|---|---|---|---|
| General Porins | 16 | Trimeric | ~600 | OmpF, OmpC (E. coli) | Moderate constriction zone, slight charge preferences |
| Specific Porins | 18 | Trimeric | Variable | LamB (maltodextrins), ScrY (sucrose) | Specialized binding sites, "greasy slide" for aromatics |
| Monomeric Porins | 14-16 | Monomeric | Variable | OmpA, OmpG | Smaller pores, some with signaling/structural roles |
| Eukaryotic Porins | 19 | Monomeric (can oligomerize) | ~1500-3000 | VDAC (mitochondria) | Voltage-gated, N-terminal α-helix regulation |
Oligomeric Assembly and Stability
Most bacterial porins form stable homotrimers in the native outer membrane, with each monomer constituting an independent translocation pathway [19] [21]. The trimeric assembly is stabilized by extensive interactions between monomer subunits, particularly through the pairing of the first β-strand of one monomer with the last β-strand of an adjacent monomer, creating a continuous β-sheet network across the trimer interface [21]. Additional stabilization is provided by surface loops, especially L2, which latch adjacent subunits together [21].
This quaternary structure confers significant thermodynamic stability, with wild-type trimeric porins exhibiting melting temperatures around 72°C—remarkably high for membrane proteins [21]. The exceptional stability of porins can be attributed to several factors: the extensive hydrogen-bonding network within the β-barrel that satisfies backbone polar groups; the precise interactions between hydrophobic barrel edges and membrane lipids; and in some porin variants, the presence of intramolecular disulfide bonds that provide additional rigidity [21]. This robustness allows porins to maintain structural integrity in the harsh extracellular environment and withstand physical and chemical stresses encountered during infection.
Functional Classification of Porin Pathways
General Diffusion Porins: Molecular Sieves of the Outer Membrane
General diffusion porins function as relatively non-selective molecular sieves that facilitate the passive diffusion of small, hydrophilic molecules according to concentration gradients [19] [20]. These porins, exemplified by OmpF and OmpC in Escherichia coli, form water-filled channels that allow the passage of various nutrients, ions, waste products, and unfortunately, many antibiotics [20] [21]. The permeability through these channels is governed primarily by physicochemical properties of the solute—including molecular size, hydrophilicity, and charge—rather than specific molecular recognition events [17].
The size exclusion limit for general porins typically falls below 600 Da, effectively preventing the passage of larger molecules while permitting rapid diffusion of smaller compounds [18]. Quantitative measurements using planar bilayer electrophysiology have demonstrated that single porin channels can achieve impressive transport rates, typically ranging from 10³ to 10⁶ molecules per second depending on solute characteristics [21]. For example, approximately 600 molecules of cephalosporin antibiotics can pass through a single OmpF monomer per second under physiological conditions [21]. This high throughput capacity ensures adequate nutrient supply while simultaneously creating a vulnerability to antibiotic penetration that bacteria must carefully regulate.
Substrate-Specific Porins: Facilitated Diffusion with Molecular Recognition
In contrast to general porins, substrate-specific channels employ sophisticated molecular recognition mechanisms to facilitate the selective uptake of particular nutrient classes [19] [21]. These specialized porins, such as maltoporin (LamB) for maltodextrins or ScrY for sucrose, incorporate specific binding sites within their channel interiors that selectively interact with target substrates [21]. This architecture enables a facilitated diffusion mechanism wherein the porin not only provides a physical passage but actively enhances the transport efficiency for cognate substrates while excluding unrelated molecules.
The structural basis for this specificity often involves strategically positioned aromatic residues that create a "greasy slide"— a continuous track of hydrophobic side chains that guides and binds specific substrates through the channel [21]. In LamB, for instance, arrays of tryptophan and tyrosine residues form stacking interactions with the glucose rings of maltooligosaccharides, significantly accelerating their diffusion rates compared to non-specific molecules of similar size [21]. This specialized transport mechanism allows bacteria to efficiently scavenge preferred nutrient sources from competitive environments, providing a selective growth advantage under nutrient-limited conditions.
Table 2: Functional Characteristics of Major Porin Types in Escherichia coli
| Porin | Type | Primary Substrates | Regulation | Role in Antibiotic Resistance |
|---|---|---|---|---|
| OmpF | General diffusion | Small hydrophilic molecules <600 Da | Osmolarity, pH, temperature | Major route for β-lactam, fluoroquinolone influx |
| OmpC | General diffusion | Small hydrophilic molecules <600 Da | Osmolarity, growth phase | Alternative route for antibiotic influx |
| LamB | Specific (maltodextrin) | Maltose, maltodextrins | Maltose regulon | Minor role; can permit antibiotic passage |
| PhoE | Specific (anion) | Phosphates, anions | Phosphate starvation | Enhanced anion antibiotic uptake |
| OmpA | Monomeric (slow porin) | Small molecules | Constitutive, stress-responsive | Mainly structural; deletion increases susceptibility |
| OmpG | Monomeric (non-specific) | Small peptides, nutrients | Stress conditions | Alternative pathway when major porins downregulated |
Methodologies for Porin Functional Characterization
Planar Lipid Bilayer Electrophysiology
The characterization of porin channel properties has been revolutionized by planar lipid bilayer electrophysiology, a technique that enables precise quantification of single-channel conductance and gating behavior [23] [22]. This method involves reconstituting purified porin proteins into artificial lipid membranes that separate two electrolyte-filled chambers, allowing researchers to measure current fluctuations associated with ion passage through individual porin channels [22].
Experimental Protocol:
- Membrane Formation: Create an artificial lipid bilayer across a small aperture (typically 100-200 μm in diameter) separating two buffered salt solutions.
- Porin Incorporation: Add purified porin proteins, solubilized in a mild detergent, to one chamber. Porin molecules spontaneously incorporate into the artificial membrane.
- Current Measurement: Apply a constant voltage across the membrane and monitor current flow with a high-gain amplifier.
- Data Analysis: Observe stepwise increases in conductance as individual porin channels insert into the bilayer. Each step corresponds to the opening of a single channel.
- Pore Characterization: Calculate pore size from conductance measurements using established theoretical models. Determine ionic selectivity by measuring reversal potentials under salt gradients.
This technique has been instrumental in establishing the size exclusion limits of various porins—approximately 1.7 nm for mitochondrial porins and 3 nm for chloroplast porins, for example [22]. Furthermore, electrophysiological studies have revealed the voltage-dependent gating behavior of certain porins, such as the voltage-dependent anion channel (VDAC) in mitochondrial membranes, which transitions between high-conductance anion-selective states at low membrane potentials and low-conductance cation-selective states at higher voltages [22].
Fluorescent Tracer Permeability Assays
Recent advances in porin research have leveraged fluorescent antibiotic analogues and other tracer molecules to investigate porin permeability in living bacterial cells [24]. This approach provides real-time information about porin function in native membranes under physiologically relevant conditions.
Experimental Protocol (2NBDG Uptake Assay):
- Bacterial Preparation: Grow bacterial cultures to mid-log phase under defined conditions appropriate for the porins being studied.
- Tracer Incubation: Add the fluorescent glucose analogue 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2NBDG) to the bacterial suspension at concentrations typically ranging from 10-100 μM.
- Uptake Monitoring: Measure fluorescence accumulation over time using flow cytometry or time-lapse fluorescence microscopy for single-cell analysis.
- Ion Modulation: Test the effects of periplasmic ions on porin permeability using ionophores (e.g., CCCP for H+, valinomycin for K+) or genetically encoded ion pumps (e.g., ArchT for targeted periplasmic acidification).
- Mutant Analysis: Compare tracer uptake between wild-type and porin-deficient strains to determine the contribution of specific porins to overall permeability.
This methodology was pivotal in demonstrating the metabolic control of porin permeability, revealing how changes in periplasmic H+ and K+ concentrations dynamically regulate porin conductance in response to metabolic status [24]. The technique offers the significant advantage of assessing porin function in situ, capturing potential regulatory interactions and compensatory mechanisms that might be absent in purified reconstitution systems.
Figure 1: Experimental Workflow for Fluorescent Tracer Permeability Assays. This diagram illustrates the key components and regulatory influences in porin permeability measurements using fluorescent tracers like 2NBDG.
Minimum Inhibitory Concentration (MIC) Profiling
Systematic analysis of antibiotic susceptibility in porin-deficient mutants provides critical information about the contribution of specific porins to antibiotic influx [20]. This approach involves constructing isogenic strains with targeted deletions in porin genes and comparing their antibiotic resistance profiles to wild-type parents.
Experimental Protocol:
- Strain Construction: Create in-frame deletion mutants of specific porin genes using λ Red recombinase-mediated homologous recombination or similar methods.
- Growth Standardization: Prepare bacterial suspensions to a standardized density (e.g., 0.5 McFarland standard, approximately 1.5 × 10⁸ CFU/mL).
- Agar Dilution Assay: Incorporate two-fold serial dilutions of antibiotics into Mueller-Hinton agar plates.
- Inoculation and Incubation: Spot 10⁴ to 10⁵ CFU of each strain onto antibiotic-containing plates and incubate at 37°C for 16-20 hours.
- MIC Determination: Identify the lowest antibiotic concentration that completely inhibits visible growth.
This systematic approach revealed distinct roles for various porins: OmpF deficiency conferred resistance to multiple β-lactam antibiotics; OmpA deletion increased susceptibility to diverse drug classes due to compromised membrane integrity; while other porins like LamB and YddB played more specialized roles in transporting specific antibiotic molecules [20]. The methodology provides functional data that directly links specific porins to antibiotic resistance phenotypes, information crucial for understanding clinical resistance mechanisms.
Porins in Antibiotic Resistance and Therapeutic Development
Porin-Mediated Resistance Mechanisms
Porins contribute to antibiotic resistance through several distinct mechanisms, with modifications in porin expression or structure representing a common adaptive strategy in Gram-negative pathogens [20] [17]. The downregulation of major porin pathways, particularly OmpF and OmpC in Enterobacteriaceae, significantly reduces the intracellular accumulation of hydrophilic antibiotics, including β-lactams, fluoroquinolones, and chloramphenicol [20]. This transcriptional regulation often occurs in response to environmental triggers, such as antibiotic exposure or oxidative stress, effectively diminishing the drug influx rate to levels that can be managed by efflux pumps or enzymatic inactivation systems.
Beyond quantitative changes in porin expression, structural modifications arising from mutations in porin-encoding genes can alter channel properties to limit antibiotic permeability while preserving essential nutrient uptake [17]. These mutations typically affect regions critical for channel function—particularly the constriction zone and loop structures—by reducing pore diameter, introducing charged residues that electrostatically repel antibiotics, or modifying gating behavior [17]. The remarkable adaptability of porins is evidenced by the low sequence identity (typically 20-30%) among porins from different bacterial species, despite conservation of the fundamental β-barrel architecture [21]. This sequence variability provides ample opportunity for evolutionary selection of resistance-conferring mutations while maintaining essential transport functions.
Metabolic Regulation of Porin Permeability
Emerging research has revealed sophisticated metabolic control mechanisms that dynamically regulate porin permeability in response to nutritional status and metabolic activity [24]. Single-cell imaging studies have demonstrated that porin conductance in Escherichia coli is controlled by changes in periplasmic H+ and K+ concentrations, which in turn are influenced by electron transport chain activity and inner membrane voltage-gated potassium channels (Kch) [24].
This ionic regulation operates through several interconnected mechanisms:
- Periplasmic Acidification: During growth on fatty acids, periplasmic pH decreases, promoting protonation of charged residues on the periplasmic surface of porins and reducing pore diameter through electrostatic effects.
- Potassium Modulation: High metabolic activity during glucose metabolism activates Kch channels, increasing periplasmic K+ concentration that enhances porin permeability, potentially to dissipate reactive oxygen species.
- Membrane Potential Coupling: Changes in inner membrane voltage correlate with porin activity, with depolarization events triggering increased porin permeability.
This metabolic regulation creates a paradigm where bacteria balance nutrient uptake needs against the vulnerability to antibiotic influx, effectively tuning outer membrane permeability according to metabolic status. This explains observed correlations between bacterial metabolic states and antibiotic susceptibility, such as the increased ciprofloxacin resistance of bacteria catabolizing lipids compared to those utilizing glucose [24].
Table 3: Research Reagent Solutions for Porin Studies
| Reagent/Category | Specific Examples | Primary Research Application | Key Function in Experimental Design |
|---|---|---|---|
| Fluorescent Tracers | 2NBDG, Bocillin FL, Hoechst | Porin permeability quantification | Monitor solute influx through porin channels in live cells |
| Ion Modulators | CCCP, valinomycin, oligomycin | Ion regulation of porin conductance | Selectively alter H+ or K+ gradients across membranes |
| Genetically Encoded Sensors | pHluorin, pHuji, GINKO1/2, QuasAr2 | Real-time ion and membrane potential monitoring | Measure dynamic changes in periplasmic/cytoplasmic ions and voltage |
| Optogenetic Tools | ArchT (light-activated proton pump) | Targeted periplasmic acidification | Precisely control periplasmic pH with temporal resolution |
| Genetic Tools | λ Red recombinase, pKD46, pCP20 | Isogenic porin mutant construction | Create targeted gene deletions and modifications for functional studies |
| Antibiotic Libraries | Diverse β-lactams, fluoroquinolones, others | MIC profiling and resistance assessment | Determine permeability coefficients and resistance contributions |
Porins as Therapeutic Targets and Adjuvants
The critical role of porins in antibiotic permeability has positioned them as attractive targets for novel therapeutic interventions against multidrug-resistant Gram-negative pathogens [18]. Two primary strategies have emerged: developing compounds that directly target porins to enhance antibiotic influx, and exploiting porin biology to improve existing antibiotics.
Polymyxin- and Aminoglycoside-Based Outer Membrane Permeabilizers: These established antibiotic classes possess inherent outer membrane-disrupting properties independent of their primary antimicrobial mechanisms [18]. Structural modification of these compounds has generated novel permeabilizers that synergize with conventional antibiotics:
- Polymyxin Derivatives: Engineered analogs with reduced intrinsic toxicity that disrupt LPS organization through electrostatic interactions with lipid A, compromising OM integrity.
- Aminoglycoside Variants: Modified structures that maintain the cationic properties necessary for OM disruption while minimizing ribosomal binding and associated toxicity.
These permeabilizers function primarily by displacing the divalent cations that bridge adjacent LPS molecules, thereby disrupting the ordered LPS lattice and increasing general permeability to hydrophobic compounds [18]. When administered in combination with conventional antibiotics, they can potentiate activity against resistant strains by enabling increased drug influx, effectively bypassing porin-related resistance mechanisms.
Figure 2: Porin-Mediated Resistance and Therapeutic Bypass Strategies. This diagram illustrates the relationship between porin-based resistance mechanisms and interventional approaches that utilize outer membrane permeabilizers.
The intricate architecture and regulated permeability of porin pathways represent a fundamental determinant of outer membrane barrier function in Gram-negative bacteria. The structural and functional distinction between general diffusion porins and substrate-specific channels establishes a sophisticated permeability network that balances nutritional requirements with protection against environmental threats, including antimicrobial agents. The emerging understanding of metabolic regulation of porin activity reveals an additional layer of complexity, demonstrating how bacteria dynamically adjust membrane permeability in response to nutritional status and metabolic demands.
From a clinical perspective, porin modifications constitute a significant resistance mechanism that diminishes the intracellular concentration of antibiotics, contributing to the escalating challenge of multidrug-resistant Gram-negative infections. The methodological advances in porin research—particularly single-cell imaging techniques and systematic genetic approaches—have unveiled the dynamic regulation and functional diversity of these channels, providing new insights into bacterial adaptation mechanisms under antibiotic pressure.
Future research directions should focus on elucidating the structural basis of porin regulation by periplasmic ions, developing high-throughput screening methods for porin-targeted permeabilizers, and exploring the therapeutic potential of manipulating porin expression or function. As our understanding of porin biology continues to evolve, so too will opportunities for innovative therapeutic strategies that exploit these essential gateway proteins to overcome antibiotic resistance in Gram-negative pathogens.
The intrinsic resistance of Gram-negative bacteria presents a formidable challenge in clinical settings, primarily mediated by the sophisticated architecture of the outer membrane (OM). This asymmetric lipid bilayer, densely packed with lipopolysaccharides (LPS) and punctuated by selective porin channels, acts as a potent permeability barrier that restricts antibiotic penetration. Combined with efflux pump systems, this native structure defines baseline antibiotic susceptibility profiles and contributes significantly to the multidrug-resistant (MDR) phenotype prevalent in pathogens such as Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii. This technical review examines the molecular determinants of OM-mediated intrinsic resistance, explores experimental methodologies for its investigation, and discusses emerging strategies to overcome this barrier for therapeutic intervention.
The outer membrane of Gram-negative bacteria represents a unique biological interface that performs the dual function of providing protection while permitting selective exchange with the environment. This asymmetrical bilayer contains phospholipids in the inner leaflet and lipopolysaccharides (LPS) in the outer leaflet, creating a chemically distinct boundary [10]. The intrinsic resistance afforded by this structure significantly limits treatment options for Gram-negative infections, with estimates indicating that more than 2.8 million antimicrobial-resistant infections occur annually in the U.S. alone, resulting in approximately 35,000 deaths each year [25].
The clinical urgency is further highlighted by the World Health Organization's classification of carbapenem-resistant Enterobacterales, Acinetobacter baumannii, and Pseudomonas aeruginosa as critical priority pathogens [26]. Understanding the molecular basis of OM-mediated resistance is therefore paramount for developing novel therapeutic strategies to overcome this pervasive clinical challenge.
Structural Foundations of the Outer Membrane
Lipopolysaccharide Architecture and Barrier Function
The LPS layer forms the primary interface between Gram-negative bacteria and their environment, creating a formidable permeability barrier. Each LPS molecule consists of three structural domains:
- Lipid A: A glucosamine-based phospholipid anchor containing six saturated fatty acid chains (compared to two in typical phospholipids) that strongly interact laterally, creating a dense, low-fluidity membrane surface [10]
- Core oligosaccharide: A branched structure containing 6-10 sugars plus two Kdo (3-deoxy-D-manno-oct-2-ulosonic acid) residues linked to lipid A
- O-antigen: A distal polysaccharide of varying length (1-40 repeating units) that contributes to strain-specific immunity and surface properties [10]
The tight packing of saturated fatty acid chains in Lipid A, combined with strong electrostatic interactions between anionic groups bridged by divalent cations (Mg²⁺ and Ca²⁺), creates a hydrophobic barrier with exceptionally low fluidity that effectively excludes many hydrophobic antibiotics [10] [26]. Mutants with truncated LPS cores ("deep rough" mutants) demonstrate significantly increased sensitivity to hydrophobic antibiotics, bile salts, and detergents, confirming the critical protective role of intact LPS structure [10].
Porin-Mediated Permeation Pathways
For hydrophilic antibiotics, the primary penetration route occurs through general diffusion porins—β-barrel proteins that form water-filled channels across the OM. In E. coli, the major porins OmpF and OmpC form trimers, with each monomer creating a distinct channel:
- OmpF: Forms larger channels (approximately 1.16 nm diameter) preferentially expressed under low osmolarity conditions
- OmpC: Forms slightly narrower channels (approximately 1.08 nm diameter) dominant under high osmolarity [27]
These porins function as molecular sieves with size exclusion limits of approximately 600 Da, effectively preventing larger molecules from entering the periplasmic space [27]. The specificity and abundance of these porins are regulated by sophisticated systems including the OmpR-EnvZ two-component system and small regulatory RNAs (sRNAs) such as MicF and MicC that fine-tune porin expression in response to environmental conditions [27].
Table 1: Major Porins in Escherichia coli and Their Regulatory Elements
| Porin | Channel Size | Expression Conditions | Primary Regulators | Permeability Characteristics |
|---|---|---|---|---|
| OmpF | ~1.16 nm | Low osmolarity, nutrient limitation | OmpR-EnvZ, MicF RNA | Larger pore, more permeable to β-lactams |
| OmpC | ~1.08 nm | High osmolarity, nutrient abundance | OmpR-EnvZ, MicC RNA | Smaller pore, restrictive barrier |
| OmpA | Variable | Constitutive | σE-dependent sRNAs | Primarily structural, limited pore activity |
Complementary Resistance Mechanisms
Efflux Pump Synergy with OM Barrier
Even when antibiotics successfully traverse the OM, Gram-negative bacteria employ a second line of defense through multidrug efflux pumps. The most clinically significant are the Resistance-Nodulation-Division (RND) family pumps, which are unique to Gram-negative bacteria [26]. These sophisticated protein complexes span the entire cell envelope:
- Inner membrane component: Energy-dependent transporter (e.g., AcrB in E. coli)
- Periplasmic adaptor: Membrane fusion protein (e.g., AcrA) that bridges the inner and outer membranes
- Outer membrane component: Channel-forming protein (e.g., TolC) that provides exit conduit to extracellular space [27]
The synergy between the OM permeability barrier and efflux systems creates a highly effective defense mechanism. The restricted penetration through the OM allows efflux pumps more time to intercept and export antibiotics, significantly reducing the intracellular concentrations achievable by many drug classes [10] [26].
Antibiotic-Inactivating Enzymes
For compounds that successfully achieve intracellular accumulation, Gram-negative bacteria possess numerous antibiotic-inactivating enzymes including:
- β-lactamases (e.g., ESBLs, KPC, NDM, OXA) that hydrolyze β-lactam rings
- Aminoglycoside-modifying enzymes (acetyltransferases, phosphotransferases, nucleotidyltransferases)
- Ribosome-modifying methyltransferases that alter drug target sites [26]
These enzymatic systems provide a third layer of defense that works in concert with permeability barriers and efflux mechanisms to establish the characteristic intrinsic resistance profile of Gram-negative pathogens.
Experimental Approaches for Investigating Intrinsic Resistance
Genome-Wide Mutant Screening
Systematic identification of genetic determinants governing intrinsic resistance has been facilitated by comprehensive mutant libraries. The Keio collection of E. coli single-gene knockouts (~3,800 strains) has enabled genome-wide screens for hypersusceptibility mutants [28] [29].
Protocol: Genome-wide susceptibility screening using the Keio collection
- Culture preparation: Grow knockout strains in 96-well format with LB media supplemented with antibiotic at IC₅₀ concentrations
- Growth assessment: Measure optical density at 600 nm after 18-24 hours incubation
- Data normalization: Express growth as fold-change compared to wild-type controls
- Hit identification: Classify strains with growth lower than two standard deviations from median as hypersensitive
- Validation: Confirm hypersusceptibility on solid media containing MIC, MIC/3, and MIC/9 antibiotic concentrations [28]
This approach identified 35 trimethoprim-hypersensitive and 57 chloramphenicol-hypersensitive mutants, with enrichment in genes involved in cell envelope biogenesis, membrane transport, and information transfer pathways [28] [29]. Particularly strong sensitization occurred with knockouts of:
- acrB (efflux pump component)
- rfaG and lpxM (LPS biosynthesis)
- nudB (folate metabolism, trimethoprim-specific) [28]
Figure 1: Experimental workflow for genome-wide identification of intrinsic resistance genes using the Keio collection of E. coli knockouts.
Transposon Sequencing (Tn-seq) for Resistance Gene Discovery
Tn-seq represents a powerful complementary approach that enables genome-wide assessment of gene contributions to antibiotic resistance under various selective conditions.
Protocol: Tn-seq for susceptibility determinant identification
- Library construction: Generate random transposon insertion mutant pools (>60,000 unique mutants)
- Selective pressure: Grow pools in sub-MIC antibiotics (typically causing 20-30% growth inhibition)
- Sample collection: Isolate DNA from start point and after 8 population doublings
- Sequencing & analysis: Amplify transposon insertion sites by PCR, sequence, and map to reference genome
- Fitness calculation: Determine gene-level fitness values based on insertion abundance changes [30]
Application in Acinetobacter baumannii identified 327 candidate susceptibility determinants, including ten genes affecting resistance to at least half of 20 tested antibiotics, highlighting both specific and broad-spectrum resistance elements [30].
Research Reagent Solutions for Intrinsic Resistance Studies
Table 2: Essential Research Tools for Investigating Outer Membrane-Mediated Resistance
| Resource/Tool | Specifications | Research Applications | Key Features |
|---|---|---|---|
| Keio Collection [28] [29] | ~3,800 single-gene E. coli K-12 knockouts | Genome-wide susceptibility screening, identification of intrinsic resistome | Arrayed format, precise gene deletions, kanamycin-marked |
| Transposon Mutant Libraries [30] | >60,000 unique Tn10 or mariner insertions | Tn-seq fitness profiling, essentiality assessment under antibiotic pressure | Pooled format, deep coverage, quantitative fitness measurements |
| CARD Database [31] | 8,582 ontology terms, 6,442 reference sequences | AMR gene annotation, resistome prediction, mutation analysis | Curated resistance ontology, RGI prediction tool, regular updates |
| Porin-Specific Antibodies [27] | Anti-OmpF, Anti-OmpC, Anti-OmpA | Porin expression quantification, localization studies | Monitor porin regulation under different conditions |
| Efflux Pump Inhibitors [28] | Chlorpromazine, PAβN, CCCP | Efflux activity assays, combination therapy studies | Chemical inhibition of RND-type pumps, synergy testing |
Therapeutic Strategies to Overcome OM-Mediated Resistance
Permeabilization Approaches
Several strategies aim to disrupt the OM barrier to enhance antibiotic penetration:
- Membrane-active agents: Polymyxin derivatives like polymyxin B nonapeptide (PMBN) compete for LPS binding sites, displacing stabilizing divalent cations and increasing membrane permeability [10]
- Chelators: EDTA and related compounds remove Ca²⁺ and Mg²⁺ ions from LPS, disrupting cross-bridging and creating phospholipid patches in the outer leaflet that are more permeable to hydrophobic compounds [10]
- Cationic peptides: Natural and synthetic peptides that target LPS and create transient disruptions in OM integrity
Efflux Pump Inhibition
Efflux pump inhibitors (EPIs) offer a complementary approach by blocking the active removal of antibiotics. However, recent research reveals limitations to this strategy:
- Genetic vs. pharmacological inhibition: While ΔacrB knockouts show dramatically reduced resistance evolution, pharmacological inhibition with chlorpromazine led to rapid resistance development against the EPI itself [28]
- Multidrug adaptation: Exposure to EPI-antibiotic combinations can select for mutations conferring broad-spectrum resistance beyond the target antibiotic [28]
Exploiting Native Transport Pathways
Structural-guided drug design aims to create antibiotics with improved penetration properties through native porin pathways. Molecular modeling of porin-antibiotic interactions enables optimization of key physicochemical parameters including:
- Molecular size/weight (preferably <600 Da)
- Hydrophilicity (favoring porin permeation)
- Charge distribution (minimizing interactions with porin constriction zones) [27] [26]
Figure 2: Strategic approaches to overcome outer membrane-mediated intrinsic resistance in Gram-negative bacteria.
Evolutionary Considerations and Resistance Development
Laboratory evolution experiments reveal that bacteria can adapt to perturbations in intrinsic resistance pathways through diverse genetic mechanisms:
- Sub-inhibitory antibiotic exposure: ΔacrB, ΔrfaG, and ΔlpxM mutants exposed to sub-MIC trimethoprim developed resistance through drug-specific mutations rather than compensatory evolution of the disrupted pathways [28]
- Differential adaptability: Resistance-conferring mutations could bypass defects in LPS biosynthesis more effectively than efflux deficiencies, suggesting efflux inhibition may provide more durable resistance prevention [28]
- Evolutionary constraints: Under high drug concentrations, intrinsic resistance knockouts were driven to extinction more frequently than wild-type strains, supporting the potential of targeting these pathways for "resistance-proofing" [28]
The native structure of the Gram-negative outer membrane constitutes a sophisticated, multifunctional barrier that defines baseline antibiotic susceptibility. Its asymmetric organization, combining a densely packed LPS leaflet with selective porin channels, creates a formidable physical and chemical obstacle to antimicrobial penetration. When combined with efflux systems and antibiotic-inactivating enzymes, this intrinsic resistome presents a challenging landscape for antibiotic development.
Advanced genetic tools and screening methodologies have enabled systematic dissection of the components comprising this resistance network, revealing both expected and unexpected genetic determinants. While strategies to disrupt this barrier show promise, evolutionary experiments highlight the remarkable adaptability of bacterial pathogens to circumvent these interventions. Future therapeutic approaches will likely require combination strategies that target multiple resistance mechanisms simultaneously while considering the evolutionary trajectories that enable resistance development. The ongoing characterization of the intrinsic resistome at molecular and structural levels continues to provide critical insights for developing next-generation antimicrobials capable of overcoming these native defense systems.
The intrinsic antibiotic resistance of Gram-negative bacteria presents a formidable challenge in clinical settings, largely due to the synergistic actions of two core defensive systems: the outer membrane (OM) permeability barrier and multidrug efflux pumps [32] [33]. Independently, each system provides a measure of protection; however, their functional interplay creates a highly effective, dual-layered defense that can profoundly reduce the intracellular concentration of antimicrobial agents [32] [33]. This synergy between passive permeability control and active efflux establishes a robust resistance phenotype that is difficult to overcome. The World Health Organization has identified antibiotic resistance as a critical global threat, with estimates attributing 4.95 million deaths to antimicrobial resistance in 2019, a figure projected to rise dramatically without effective interventions [32]. Understanding the molecular details of this collaborative defense is not merely an academic exercise but an urgent necessity to inform the development of novel therapeutic strategies, including efflux pump inhibitors and OM permeabilizers, aimed at resensitizing resistant pathogens [34].
Fundamental Mechanisms of the Outer Membrane Barrier
Structural Organization and Composition
The Gram-negative outer membrane is a unique asymmetric bilayer that serves as a formidable physical barrier. Its outer leaflet is predominantly composed of lipopolysaccharide (LPS), which confers rigidity and a strong negative surface charge, while the inner leaflet consists of phospholipids [32] [5] [4]. A typical LPS molecule contains three structural domains: lipid A (a glucosamine-based phospholipid anchoring the molecule to the membrane), a core oligosaccharide, and the distal O-antigen polysaccharide [5] [4]. This specialized organization is crucial to the membrane's barrier function. The LPS layer forms a tightly packed, gel-like mesh that is reinforced by divalent cations (Mg²⁺ and Ca²⁺) that bridge adjacent LPS molecules through electrostatic interactions [32] [5]. This structure severely restricts the penetration of hydrophobic compounds and provides an exceptional level of innate resistance to many antimicrobial agents.
Pathways for Antibiotic Permeation
Antibiotics and other solutes primarily traverse the outer membrane through two distinct pathways, each with specific physicochemical requirements:
- Porin-mediated pathway: General diffusion porins such as OmpF, OmpC, and OprD form water-filled channels that allow the passive diffusion of small, hydrophilic antibiotics like β-lactams, fluoroquinolones, and aminoglycosides [3] [5] [4]. These porins typically exhibit size exclusion limits, often permitting passage of molecules up to approximately 600 Da [4].
- Lipid-mediated pathway: Hydrophobic antibiotics such as macrolides and rifamycins traverse the OM by passive diffusion through the lipid bilayer itself [3] [5] [4]. Their permeation depends on physicochemical properties including lipophilicity, molecular size, and polarity [32].
Table 1: Major Outer Membrane Porins and Their Antibiotic Substrates
| Porin | Bacterial Species | Antibiotic Substrates |
|---|---|---|
| OmpF/OmpC | Escherichia coli | β-lactams, fluoroquinolones, tetracyclines, chloramphenicol [33] [4] |
| OprD | Pseudomonas aeruginosa | Carbapenems (imipenem, meropenem) [33] |
| OmpK36/OmpK35 | Klebsiella pneumoniae | β-lactams, fluoroquinolones [4] |
| Omp36 | Enterobacter aerogenes | Cephalosporins, carbapenems [4] |
Multidrug Efflux Pump Systems
Architectural Diversity and Energy Coupling
Efflux pumps are active transporters that expel toxic compounds, including antibiotics, from the bacterial cell. They are classified into five major superfamilies based on their structure and energy source [34] [35] [36]:
- Resistance-Nodulation-Division (RND): Proton motive force-dependent; often form tripartite complexes spanning both inner and outer membranes (e.g., AcrAB-TolC in E. coli, MexAB-OprM in P. aeruginosa) [34] [35].
- Major Facilitator Superfamily (MFS): Proton motive force-dependent; primarily found in Gram-positive bacteria but present in Gram-negatives [34] [35].
- ATP-Binding Cassette (ABC): ATP hydrolysis-dependent; the only primary active transporters among efflux pumps [35] [36].
- Small Multidrug Resistance (SMR): Proton motive force-dependent; compact size with broad substrate recognition [34] [35].
- Multidrug and Toxic Compound Extrusion (MATE): Sodium or proton motive force-dependent [34].
Of these, RND-type pumps are particularly significant in Gram-negative pathogens due to their broad substrate specificity and contribution to clinical multidrug resistance [34].
Physiological Functions Beyond Antibiotic Resistance
While efflux pumps are widely recognized for their role in antibiotic resistance, their fundamental physiological functions predate clinical antibiotic use [34] [35]. These natural roles include:
- Removal of bile acids and fatty acids in intestinal pathogens (e.g., AcrAB in E. coli) [35]
- Export of virulence factors and bacterial metabolites [34]
- Heavy metal detoxification and stress response modulation [34] [35]
- Biofilm formation and quorum sensing signal transport [35]
- Transport of cell envelope components [34]
The ability of these pumps to recognize antibiotics likely represents an accidental exploitation of their inherent capacity to identify molecules based on general physicochemical properties such as hydrophobicity, aromaticity, and ionizable character, rather than specific molecular structures [35].
Table 2: Major RND Efflux Pumps in Multidrug-Resistant Pathogens
| Efflux Pump | Bacterial Species | Regulator | Antibiotic Substrates |
|---|---|---|---|
| AdeABC | Acinetobacter baumannii | AdeRS, BaeSR | Aminoglycosides, β-lactams, tetracyclines, fluoroquinolones, tigecycline* [34] |
| MexAB-OprM | Pseudomonas aeruginosa | MexR | β-lactams, fluoroquinolones, chloramphenicol, tetracyclines, macrolides [33] |
| AcrAB-TolC | Escherichia coli | AcrR, MarA, SoxS | β-lactams, fluoroquinolones, chloramphenicol, tetracyclines, oxazolidinones [33] [35] |
| MexXY-OprM | Pseudomonas aeruginosa | MexZ | Aminoglycosides, macrolides, tetracyclines, fluoroquinolones [33] |
Experimental Approaches for Studying Permeability and Efflux
Quantitative Assessment of Antibiotic Accumulation
Direct measurement of intracellular antibiotic accumulation provides crucial insights into the efficiency of compound penetration and efflux. Liquid chromatography-mass spectrometry (LC-MS) enables simultaneous quantitation of multiple antibiotics within bacterial cells [37].
Protocol: LC-MS-based Antibiotic Accumulation Assay
- Bacterial culture: Grow bacterial strains of interest to mid-log phase (OD₆₀₀ ≈ 0.5-0.6) in appropriate medium.
- Antibiotic exposure: Incubate bacteria with therapeutic concentrations of target antibiotics (e.g., 10× MIC) for 4 hours at 37°C with shaking.
- Sample processing: Harvest cells by rapid centrifugation (10,000 × g, 2 min, 4°C), wash twice with ice-cold phosphate-buffered saline to remove extracellular antibiotic.
- Cell lysis: Lyse cell pellets using mechanical disruption (bead beating) or chemical lysis in acetonitrile:water (1:1) with 0.1% formic acid.
- LC-MS analysis: Separate antibiotics using reverse-phase chromatography (C18 column) with gradient elution (water:acetonitrile + 0.1% formic acid). Perform detection using a high-resolution mass spectrometer in multiple reaction monitoring (MRM) mode.
- Data analysis: Normalize antibiotic signals to total cellular protein and compare to standard curves for absolute quantitation [37].
This approach has revealed striking variations in antibiotic accumulation in Mycobacterium abscessus, with greater than 1000-fold differences between the highest and lowest accumulating compounds, and a significant negative correlation between intracellular accumulation and MIC for drugs with intracellular targets [37].
Minimum Inhibitory Concentration (MIC) Reduction Assays
Evaluating the potentiation effects of OM-disrupting agents provides functional evidence of synergistic interactions between permeability barriers and efflux systems.
Protocol: MIC Reduction Assay with OM Permeabilizers
- Bacterial preparation: Prepare standardized bacterial inoculum (∼5×10⁵ CFU/mL) from mid-log phase cultures.
- Permeabilizer selection: Utilize OM-disrupting agents from different mechanistic classes:
- Cationic peptides: Colistin (0.35 μM) displaces divalent cation bridges between LPS molecules [32]
- Cationic steroids: Squalamine (5 μM) integrates into OM via electrostatic interactions [32]
- Chelators: EDTA (1 mM) extracts Ca²⁺ and Mg²⁺ ions that stabilize LPS [32]
- Polyaminoisoprenyl derivatives: NV716 (10 μM) binds to LPS and induces OM destabilization [32]
- MIC determination: Perform broth microdilution following CLSI guidelines with antibiotic serial dilutions in the presence and absence of sub-inhibitory concentrations of permeabilizers.
- Data interpretation: Consider a ≥4-fold reduction in MIC in the presence of permeabilizer as indicative of significant potentiation [32].
This methodology has demonstrated dramatic potentiation effects, such as a 128-fold reduction in doxycycline MIC against P. aeruginosa when combined with NV716 [32].
Computational Approaches for Studying Permeation
Molecular dynamics (MD) simulations provide atomic-level insights into antibiotic permeation across the OM. These computational methods complement empirical approaches by modeling molecular interactions between antibiotics and membrane components.
Protocol: Molecular Dynamics Simulation of OM Permeation
- Membrane modeling: Construct an asymmetric OM bilayer with LPS in the outer leaflet and phospholipids in the inner leaflet, incorporating relevant ion concentrations (Ca²⁺, Mg²⁺) to stabilize LPS organization [33] [38].
- System setup: Solvate the membrane in explicit water, add physiological salt concentration (0.15 M NaCl), and neutralize system charge.
- Simulation parameters: Use force fields (e.g., CHARMM36, GROMOS) parameterized for membranes and carbohydrates. Apply periodic boundary conditions and maintain constant temperature (310 K) and pressure (1 atm).
- Permeation analysis: Calculate position-dependent free energy profiles (potential of mean force) using umbrella sampling or metadynamics to determine thermodynamic barriers to permeation [33].
- Machine learning integration: Train models on chemical fragment libraries to identify molecular features associated with efficient OM permeation [33].
Recent MD simulations of P. aeruginosa OM have revealed that the LPS layer exhibits slow lateral diffusion resembling lipids in a gel state, creating both thermodynamic and kinetic barriers to antibiotic penetration [33] [38].
The Synergistic Defense Model
Kinetic Interplay Between Influx and Efflux
The synergistic relationship between the OM permeability barrier and efflux pumps creates a highly efficient defense system governed by the kinetic balance between passive influx and active efflux [32] [33]. The OM serves as a primary filter that significantly reduces the rate of antibiotic entry, while efflux pumps actively remove molecules that successfully traverse the OM before they reach their intracellular targets [32]. This collaboration is particularly effective because even minor reductions in influx or modest increases in efflux can profoundly impact net intracellular accumulation due to their multiplicative rather than additive effects [32] [33]. Recent kinetic models demonstrate how subtle perturbations in this balance can restore bacterial susceptibility to previously ineffective antibiotics [32] [33].
Quantitative Evidence of Synergy
Experimental data from combination studies with OM permeabilizers and antibiotics provide compelling evidence for this synergistic relationship. Research on P. aeruginosa demonstrates that disruption of the OM permeability barrier dramatically enhances the activity of antibiotics that are typically ineffective against this pathogen due to limited penetration [32].
Table 3: Potentiation of Antibiotic Activity by OM Permeabilizers in P. aeruginosa
| Antibiotic Class | Antibiotic | Baseline MIC (mg/L) | MIC with NV716 (10 µM) | Fold Reduction | MIC with EDTA (1 mM) | Fold Reduction |
|---|---|---|---|---|---|---|
| Tetracyclines | Doxycycline | 64 | 0.5 | 128× | 1 | 64× |
| Tetracyclines | Minocycline | 32 | 1 | 32× | 2 | 16× |
| Amphenicols | Chloramphenicol | 64 | 4 | 16× | 4 | 16× |
| Amphenicols | Florfenicol | 256 | 4 | 64× | 16 | 16× |
| Macrolides | Azithromycin | 128 | 32 | 4× | 64 | 2× |
| Rifamycins | Rifampicin | 128 | 32 | 4× | 64 | 2× |
Beyond simple MIC reductions, mechanistic studies reveal that the physicochemical properties of antibiotics—including molecular size, lipophilicity, polarizability, and polar surface area—collectively determine their susceptibility to this synergistic defense [32]. No single parameter reliably predicts potentiation potential; instead, these factors operate within a multidimensional "responsive zone" where optimal ranges of size, polarity, and lipophilicity act synergistically to enhance antibiotic uptake when OM disruption occurs [32].
Research Toolkit: Essential Reagents and Methodologies
Table 4: Research Reagent Solutions for Studying OM Permeability and Efflux
| Reagent/Method | Function/Application | Key Examples |
|---|---|---|
| OM Permeabilizers | Disrupt OM integrity to study permeability contributions to resistance | EDTA (chelator), colistin (cationic peptide), NV716 (LPS binder), squalamine (aminosterol) [32] |
| Efflux Pump Inhibitors | Block efflux activity to assess pump contributions to resistance | CCCP (proton motive force disruptor), PABN (RND pump inhibitor), verapamil (P-glycoprotein inhibitor) [34] [35] |
| Genomic Tools | Identify and characterize efflux systems and OM modifications | Transposon mutagenesis screens, knockout mutants, overexpression strains [34] [37] |
| Analytical Techniques | Quantify antibiotic accumulation and localization | LC-MS/MS for intracellular antibiotic measurement, fluorescent substrate tracking [37] |
| Computational Approaches | Model molecular interactions and permeation pathways | Molecular dynamics simulations of OM, machine learning for permeability prediction [33] [38] |
| Porin Modulators | Study porin-mediated uptake pathways | Antibiotics with known porin dependencies (β-lactams, fluoroquinolones) [3] [4] |
Therapeutic Implications and Future Perspectives
The interplay between OM permeability and efflux pumps presents both challenges and opportunities for antibiotic development. Strategic approaches to counter this synergistic defense include:
Combination therapies with OM-disrupting agents: Co-administration of antibiotics with sub-inhibitory concentrations of OM permeabilizers can dramatically enhance activity against Gram-negative pathogens [32]. The dramatic potentiation of tetracyclines and amphenicols by NV716 and EDTA demonstrates the therapeutic potential of this approach [32].
Efflux pump inhibitors (EPIs): Developing compounds that inhibit multidrug efflux pumps could resensitize resistant strains to existing antibiotics [34] [35]. While no EPIs have yet reached clinical use, natural products including carotenoids, flavonoids, and alkaloids show promise as starting points for development [35].
Physicochemical optimization of antibiotics: Designing compounds with properties that favor OM permeation while minimizing efflux recognition represents a rational approach to overcoming bacterial defenses [32] [33]. The multidimensional "responsive zone" concept, which considers size, polarity, and lipophilicity collectively, provides a framework for such optimization [32].
The continuing emergence of multidrug-resistant Gram-negative pathogens underscores the urgent need to fully understand and develop countermeasures against their sophisticated defensive systems. Future research should focus on elucidating the precise molecular mechanisms of synergistic defense, developing advanced models that integrate both OM permeability and efflux activities, and translating these insights into novel therapeutic strategies that can restore the efficacy of existing antibiotics against even the most recalcitrant pathogens.
Breaching the Wall: Strategies to Enhance Antibiotic Penetration
The outer membrane (OM) of Gram-negative bacteria presents a formidable barrier that protects the cell from external threats, including antibiotics [10]. This membrane's asymmetric structure, composed of phospholipids in the inner leaflet and lipopolysaccharides (LPS) in the outer leaflet, creates a highly impermeable surface that significantly contributes to intrinsic antibiotic resistance [10] [39]. Porins, β-barrel transmembrane proteins that form water-filled channels across the OM, serve as the primary gatekeepers for hydrophilic compound permeation, including many clinically important antibiotics such as β-lactams, fluoroquinolones, and carbapenems [40] [41]. The molecular rules governing permeation through these porins are therefore critical for understanding antibiotic resistance and designing effective antimicrobial agents against Gram-negative pathogens.
The challenge of molecular permeation through the OM is exemplified by the high failure rates in antibiotic discovery programs. Most hits identified in high-throughput screening campaigns fail to progress to lead compounds due to poor molecular uptake or intracellular accumulation [40] [41]. Unlike standard druglikeness rules such as the Lipinski Rule of 5, permeation through the bacterial OM does not follow conventional medicinal chemistry principles, necessitating a specialized understanding of the molecular descriptors that govern porin permeability [41]. This technical guide synthesizes current research to provide a comprehensive framework for understanding and applying porin permeation rules, with a specific focus on optimizing molecular size, charge, and polarity to enhance antibiotic influx.
Molecular Descriptors Governing Porin Permeation
Fundamental Physicochemical Properties
The permeation of molecules through porin channels is governed by a complex interplay of steric (size-related) and electrostatic (charge-related) factors. Research analyzing the correlation between whole-cell compound accumulation in Escherichia coli and predicted porin permeability coefficients has revealed a strong linear relationship (R = 0.74), confirming the critical role of porins in compound uptake [40] [41]. The key molecular descriptors identified through these studies can be categorized into steric/structural properties and electrostatic properties, each contributing differently to the permeation process.
Table 1: Key Molecular Descriptors for Porin Permeation
| Molecular Descriptor | Impact on Permeation | Optimal Range/Characteristic | Experimental Support |
|---|---|---|---|
| Net Charge | Positive charge enhances permeation through cation-selective porins | +1 to +2 | Strong correlation with accumulation in E. coli [40] [41] |
| Minimal Projection Area | Determines steric feasibility of passage | <80 Ų (smaller generally better) | Critical for initial entry into porin constriction zone [40] |
| Transversal Dipole Moment | Favors alignment with porin's internal electric field | >10 Debye | Correlates with improved accumulation [40] [41] |
| Total Dipole Moment | Influences interaction with porin electrostatics | >10 Debye | Distinguishes good from bad accumulators [40] |
| Molecular Flexibility | Affects entropic penalty during permeation | Moderate flexibility ideal | Rigid molecules with low dipole show poor permeation [40] |
Analysis of accumulation data for approximately 200 molecules in E. coli revealed that molecules with electrostatic neutrality or negative charge typically function as bad accumulators, which aligns with the cation selectivity of major porins OmpF and OmpC [40] [41]. The internal negative electrostatic potential within these porins creates a preference for positively charged molecules. However, positive charge alone does not guarantee successful accumulation, as evidenced by numerous positively charged molecules falling into the bad accumulator category, highlighting the multifactorial nature of the permeation process [40].
Advanced Mechanistic Descriptors from Computational Studies
Recent computational approaches have identified additional mechanistic descriptors that provide deeper insights into the permeation process. Studies on Pseudomonas aeruginosa have evaluated 174 molecular descriptors and their correlations with antibacterial activity, identifying specific interactions between compounds and the OM as critical determinants of permeation [39]. These include:
- Interaction energy with OM environment (Δh): Quantifies the energy of interaction between the compound and various regions of the outer membrane
- Hydrogen bonding with surrounding environment (HB): Measures the number and stability of hydrogen bonds formed during translocation
- Molecular lateral displacement (Δxy): Captures the compound's movement within the membrane environment
- Molecular entropy (Δs): Reflects the conformational flexibility and ordering during permeation
These descriptors, particularly those quantifying interactions with the LPS lipid-A and oligosaccharide core sub-regions of the OM, show high correlations with permeation and growth inhibition when combined with traditional properties like hydrophobic surface area and Randic index [39].
Experimental Methods for Assessing Porin Permeation
Liposome Swelling Assays
The liposome swelling assay (LSA) represents a cornerstone method for experimental determination of porin permeability. This technique measures the relative permeability coefficients (RPCs) of compounds through purified porins reconstituted into liposomes, providing a cell-free system for direct permeability assessment [42].
Protocol Overview:
- Porin Purification: Isolate native or recombinant porins (e.g., OmpF, OmpC) from bacterial cultures using detergent extraction and chromatographic purification
- Liposome Reconstitution: Incorporate purified porins into liposomes composed of appropriate lipid mixtures that mimic the bacterial outer membrane environment
- Osmotic Swelling Measurement: Monitor liposome swelling via light scattering or microscopy when exposed to test compounds in hypo-osmotic conditions
- Data Analysis: Calculate RPC values relative to a reference compound (typically glycine) based on swelling kinetics
This method has been successfully applied to a diverse set of 41 substrates including antibiotics (amphenicols, β-lactams, fluoroquinolones) and non-antibiotic substances (amino acids, carbohydrates, nucleosides), revealing that amino acids are generally the best permeators while nucleosides and nucleotides are among the worst [42]. The assay allows for systematic evaluation of how specific molecular transformations affect permeation rates, providing critical structure-permeability relationships.
Whole-Cell Accumulation Assays
Whole-cell accumulation assays measure the intracellular concentration of compounds in bacterial cells, providing a comprehensive view of net uptake that encompasses both influx and efflux processes [40] [41].
Protocol Overview:
- Bacterial Culture: Grow standardized cultures of target bacterial strains (e.g., E. coli MG1655 or specific mutants)
- Compound Exposure: Incubate bacteria with test compounds under controlled conditions (concentration, time, temperature)
- Cell Harvesting and Washing: Rapidly separate cells from extracellular compound via filtration or centrifugation with appropriate washing
- Compound Extraction and Quantification: Lyse cells and quantify intracellular compound concentrations using HPLC-MS, fluorescence, or radioactivity detection
- Data Normalization: Normalize accumulation values to cell number (CFU) or protein content
This approach was instrumental in establishing the correlation between porin permeability and whole-cell accumulation, with studies analyzing approximately 200 molecules demonstrating that 74% of the variation in accumulation could be explained by porin permeability alone [40] [41]. The method is particularly valuable for validating computational predictions and establishing structure-accumulation relationships.
Computational and Brownian Dynamics Approaches
Computational methods have emerged as powerful tools for predicting porin permeability, enabling rapid screening of compound libraries without the need for synthesis and experimental testing.
Brownian Dynamics Protocol [43]:
- Molecular Preparation: Generate 3D structures of test compounds and energy minimize using molecular mechanics force fields
- Porin Structure Preparation: Obtain high-resolution crystal structures of target porins (e.g., OmpF, OmpC) and prepare for simulation
- Temperature-Accelerated Sampling: Employ enhanced sampling techniques to overcome translocation energy barriers
- Permeability Calculation: Estimate permeability coefficients using the inhomogeneous solubility diffusion model from trajectory analysis
- Validation: Compare computational predictions with experimental liposome swelling or accumulation data
This approach achieves approximately 14-fold acceleration compared to all-atom molecular dynamics methods while maintaining good correlation with experimental permeation rates, making it suitable for high-throughput screening of compound libraries [43]. The method enables researchers to assess the effect of different molecular transformations on permeation rates prior to chemical synthesis.
Visualization of Porin Permeation Pathways and Experimental Workflows
Diagram 1: Compound Permeation Pathways Across Gram-Negative Bacterial Membranes. This diagram illustrates the three primary routes for compound entry across the outer membrane and the competing efflux processes that reduce intracellular accumulation.
Diagram 2: Integrated Workflow for Porin Permeability Assessment. This flowchart outlines the complementary computational and experimental approaches for establishing porin permeation rules, culminating in predictive models for compound design.
The Scientist's Toolkit: Essential Research Reagents and Methods
Table 2: Key Research Reagent Solutions for Porin Permeation Studies
| Reagent/Method | Function | Application Context |
|---|---|---|
| Fluorescent Glucose Analog (2NBDG) | Tracer for porin-mediated uptake | Real-time monitoring of porin permeability in live cells [24] |
| Bocillin FL | Fluorescent penicillin analog | Specific tracking of β-lactam permeation through porins [24] |
| pHuji & pHluorin | Genetically encoded pH sensors | Monitoring periplasmic and cytoplasmic pH changes during ion flux [24] |
| GINKO1 & GINKO2 | Genetically encoded potassium sensors | Measuring cytoplasmic and periplasmic K+ concentrations [24] |
| QuasAr2 | Genetically encoded voltage sensor | Monitoring inner membrane potential changes [24] |
| ArchT Proton Pump | Optogenetic periplasmic acidification | Controlled manipulation of periplasmic pH to study porin gating [24] |
| Ionophores (CCCP, Valinomycin) | Selective ion transport across membranes | Dissecting contributions of H+ and K+ to porin regulation [24] |
| Liposome Swelling Assay Kit | In vitro permeability measurement | Cell-free assessment of compound permeation through purified porins [42] |
Metabolic Regulation of Porin Permeability
Recent research has revealed that porin permeability is not static but dynamically regulated by metabolic processes and ion gradients. Studies using single-cell imaging have demonstrated that porin permeability in E. coli is controlled by changes in periplasmic H+ and K+ concentrations [24]. This metabolic control occurs through several mechanisms:
- Periplasmic pH Regulation: Under starvation conditions, low periplasmic H+ concentration increases conductance through porins, promoting nutrient uptake. Conversely, periplasmic acidification during growth on lipid media decreases porin permeability to limit proton loss [24]
- Potassium Channel Modulation: High metabolic activity during glucose metabolism activates the inner membrane voltage-gated potassium channel Kch, increasing periplasmic potassium concentration and enhancing porin permeability, potentially to dissipate reactive oxygen species [24]
- Membrane Potential Coupling: Membrane depolarization events correlate with increased porin permeability, creating a feedback loop that links bacterial metabolic state to outer membrane permeability [24]
This metabolic regulation explains observed variations in antibiotic resistance based on bacterial metabolic state, such as increased ciprofloxacin resistance in bacteria catabolizing lipids, and identifies ion channels like Kch as potential therapeutic targets to improve bacterial killing by antibiotics [24].
Pathogen-Specific Permeation Considerations
While general principles govern porin permeation across Gram-negative bacteria, pathogen-specific differences necessitate tailored approaches for antibiotic design.
Pseudomonas aeruginosa Considerations
P. aeruginosa presents a particularly challenging permeability barrier due to its unique OM composition [39]. The OM of P. aeruginosa features:
- Highly anionic LPS molecules tightly complexed with divalent cations
- Long carbohydrate-enriched regions that shield membrane exposure
- Repellent phosphates and ionizable groups that hinder hydrophobic molecule penetration
- Strong coordination among LPS molecules via divalent cation sheets
Computational studies analyzing 1260 antimicrobial compounds against P. aeruginosa identified that descriptors quantifying compound interactions with the LPS lipid-A and oligosaccharide core sub-regions of the OM showed the highest correlations with permeation and growth inhibition [39]. These findings highlight the need for pathogen-specific permeation rules rather than universal guidelines.
Impact of LPS Structure on Permeability
The structure of lipopolysaccharides, particularly the O-antigen polysaccharide length, significantly impacts outer membrane permeability. Recent studies have revealed that:
- Long O-antigen polysaccharides compromise OM barrier integrity, increasing antibiotic susceptibility
- Balanced production of long and short LPS forms maintains OM antibiotic barrier integrity
- O-antigen transport and assembly onto the cell surface inherently weakens the OM barrier [14]
This creates a trade-off for bacterial cells between protection from host assaults and maintaining OM integrity, with important implications for antibiotic sensitivity across different bacterial strains and growth conditions [14].
The comprehensive understanding of porin permeation rules provides a robust framework for optimizing antibiotic compounds against Gram-negative pathogens. The integration of computational prediction methods with experimental validation enables rational design of compounds with enhanced permeation properties. Key principles emerge for antibiotic optimization:
- Charge Optimization: Incorporate positive charges while balancing overall molecular properties to enhance permeation through cation-selective porins
- Size and Shape Engineering: Design compounds with minimal projection areas compatible with porin constriction zones while maintaining pharmacological properties
- Dipole Moment Considerations: Optimize transversal and total dipole moments to favor alignment with porin electrostatic landscapes
- Metabolic Context Awareness: Account for pathogen-specific metabolic environments that may dynamically regulate porin permeability
- Pathogen-Specific Tailoring: Adapt molecular design to address unique permeability barriers in priority pathogens like P. aeruginosa
As antibiotic resistance continues to threaten modern medicine, the strategic application of these porin permeation rules offers a pathway to revitalize Gram-negative antibiotic discovery and combat multidrug-resistant infections.
The outer membrane of Gram-negative bacteria presents a formidable permeability barrier that significantly reduces antibiotic access to intracellular targets, rendering these pathogens inherently less susceptible to many antimicrobial agents than their Gram-positive counterparts [44]. This structural defense, combined with the escalating crisis of antimicrobial resistance (AMR), necessitates innovative therapeutic approaches that circumvent conventional penetration pathways. The World Health Organization has classified several Gram-negative bacteria, including Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae such as Escherichia coli and Klebsiella pneumoniae, as critical priorities for novel antibiotic development due to their extensive drug resistance profiles [45] [46].
Iron acquisition systems represent a critical vulnerability in bacterial physiology that can be exploited for therapeutic gain. Under iron-restricted conditions, which are actively maintained by host nutritional immunity, bacteria synthesize and secrete siderophores—small molecular iron chelators with exceptionally high affinity for Fe³⁺ [47] [48]. This review examines the strategic hijacking of these active iron transport systems through siderophore mimicry and "Trojan horse" conjugates, an approach that leverages the bacteria's own essential nutrient uptake machinery to deliver antibiotic payloads across the otherwise impermeable outer membrane barrier.
Bacterial Iron Acquisition and Siderophore Biology
The Iron Restriction Landscape
Iron is an essential element for nearly all bacterial cellular processes, serving as a cofactor for enzymes involved in electron transport, DNA synthesis, and energy metabolism [47]. However, the bioavailability of iron is severely restricted in physiological environments. At neutral pH and in the presence of oxygen, iron exists predominantly as insoluble ferric oxyhydroxides, with a free concentration of approximately 10⁻¹⁸ M—far below the 10⁻⁶ M required to support bacterial growth [49]. mammalian hosts exacerbate this scarcity through nutritional immunity, a defense mechanism that further sequesters iron via proteins like transferrin and lactoferrin, reducing serum free iron concentrations to approximately 10⁻²⁴ M during infection [47] [48].
Siderophore Structural Diversity and Uptake Mechanisms
To overcome iron restriction, bacteria produce siderophores, which are small (200-2000 Da) secondary metabolites with dedicated iron-chelating functionalities [50] [49]. Over 500 structurally distinct siderophores have been identified, categorized primarily by their iron-coordinating motifs: catecholates, hydroxamates, phenolates, carboxylates, and α-hydroxy carboxylates [50] [49]. Many siderophores incorporate mixed chelating motifs, enhancing their versatility and affinity for iron.
The bacterial iron acquisition process follows a meticulously coordinated pathway:
- Synthesis and Secretion: Bacteria biosynthesize siderophores through dedicated pathways, often encoded by iron-regulated gene clusters, and secrete them into the extracellular environment [48].
- Iron Chelation: Siderophores solubilize and chelate environmental Fe³⁺ with remarkable affinity, characterized by pFe³⁺ values ranging from 20.0 to 35.5 [48].
- Active Transport: The ferri-siderophore complexes are recognized by specific outer membrane receptors and actively transported into the cell.
Table: Major Siderophore Classes and Representative Examples
| Siderophore Class | Iron-Binding Motif | Representative Example | Producing Organism |
|---|---|---|---|
| Catecholate | Catechol | Enterobactin | Escherichia coli |
| Hydroxamate | Hydroxamic acid | Ferrichrome | Ustilago sphaerogena |
| Mixed-type | Catechol/Hydroxamate | Pyoverdine | Pseudomonas aeruginosa |
| Carboxylate | α-Hydroxy carboxylate | Rhizobactin | Sinorhizobium meliloti |
The transport machinery differs between Gram-negative and Gram-positive bacteria due to fundamental differences in cell envelope structure. In Gram-negative bacteria, the process involves:
- TonB-Dependent Transporters (TBDTs): Outer membrane proteins that recognize specific ferri-siderophore complexes [45] [50].
- TonB-ExbB-ExbD Complex: Transduces proton motive force from the inner membrane to energize transport across the outer membrane [50].
- Periplasmic Binding Proteins and ABC Transporters: Facilitate translocation across the periplasm and inner membrane [48].
Once internalized, iron is released from siderophores through enzymatic degradation or reductive processes, as siderophores have significantly lower affinity for Fe²⁺ [48].
Diagram: Siderophore and Sideromycin Uptake Pathway in Gram-Negative Bacteria. The diagram illustrates the active transport of natural siderophores and synthetic sideromycins across the Gram-negative cell envelope, highlighting the TonB-dependent outer membrane transport and ABC transporter-mediated inner membrane crossing.
The Trojan Horse Strategy: Siderophore-Antibiotic Conjugates
Conceptual Framework and Historical Precedents
The "Trojan horse" strategy involves covalently linking antibiotic agents to siderophores or siderophore-mimetic compounds to exploit bacterial iron acquisition systems for targeted drug delivery [51] [50]. This approach provides a multifaceted advantage: (1) enhanced permeation across the outer membrane through active transport rather than passive diffusion, (2) increased intracellular antibiotic accumulation, and (3) potential bypassing of efflux pump-mediated resistance [50] [49].
Nature provides precedent for this strategy in the form of sideromycins—natural siderophore-antibiotic conjugates produced by certain microorganisms as competitive weapons [44] [49]. The most extensively studied sideromycin is albomycin, produced by Streptomyces griseus, which consists of a ferrichrome-like iron-chelating moiety linked to a thioribosyl pyrimidine antibiotic that inhibits seryl-tRNA synthetase [50] [49]. Remarkably, albomycin demonstrated potent activity against both Gram-positive and Gram-negative bacteria and was clinically used in the Soviet Union as early as the 1950s [49].
Design Principles for Synthetic Conjugates
The rational design of synthetic siderophore-antibiotic conjugates (SACs) requires careful consideration of three fundamental components:
- Siderophore/Mimetic Component: Dictates receptor specificity and transport efficiency. Catechol-based motifs have emerged as particularly effective due to their recognition by multiple TBDTs [45] [49].
- Antibiotic Warhead: Determines the intracellular mechanism of action. β-Lactams have been extensively utilized due to their periplasmic targets and synthetic accessibility [45] [49].
- Linker Chemistry: Controls stability, cleavage mechanism, and release kinetics of the active antibiotic. Optimal linkers must remain stable during transit but allow efficient intracellular release [50] [49].
Table: Siderophore-Antibiotic Conjugates and Their Antibacterial Activity
| Conjugate Name | Siderophore Type | Antibiotic Warhead | Target Bacteria | Fold-Improvement in MIC* |
|---|---|---|---|---|
| Cefiderocol | Catechol | Cephalosporin | Gram-negative bacilli | >1000-fold vs. cephalosporins |
| BAMP | Bis-catechol | Ampicillin | E. coli, K. pneumoniae | Up to 8000-fold [45] |
| BLOR | Bis-catechol | Loracarbef | E. coli, K. pneumoniae | >8192-fold [45] |
| MCEF | Mixed catechol-hydroxamate | Cefaclor | A. baumannii | 8-fold [45] |
| Albomycin (natural) | Trihydroxamate | Thioribosyl pyrimidine | Gram-positive and Gram-negative | 100-fold vs. ampicillin [49] |
Fold-improvement compared to unconjugated antibiotic under iron-depleted conditions
The recent FDA approval of cefiderocol, a cephalosporin conjugated to a catechol siderophore mimetic, validates this approach clinically. Cefiderocol demonstrates enhanced potency against multidrug-resistant Gram-negative pathogens, including carbapenem-resistant strains, by exploiting iron transport systems for periplasmic delivery [49].
Quantitative Analysis of Enhanced Efficacy
Permeation Advantages and Uptake Kinetics
Siderophore conjugation dramatically enhances antibiotic permeation across the Gram-negative outer membrane through several mechanisms:
- Receptor-Mediated Active Transport: SACs bypass the size-restrictive porin channels (typically <600 Da) by utilizing the larger TBDT pores, enabling entry of larger molecular constructs [45].
- Energy-Dependent Accumulation: Unlike passive diffusion, TonB-dependent transport is energy-coupled, allowing SAC accumulation against concentration gradients [44] [50].
- Exploitation of Nutritional Stress Response: Under iron-limited conditions, TBDT expression is significantly upregulated, creating more entry points for SACs [45].
Recent studies with β-lactam-based sideromycins demonstrate extraordinary enhancements in antibacterial potency, with some conjugates showing >8000-fold improvement in minimum inhibitory concentrations (MICs) compared to their unconjugated counterparts [45]. The magnitude of enhancement is species-dependent and influenced by multiple factors, including TBDT expression profiles, endogenous siderophore competition, and iron availability.
Mechanisms Underlying Enhanced Bacterial Inhibition
The dramatically improved efficacy of SACs cannot be attributed solely to enhanced membrane permeation. Comparative studies reveal complex, species-specific mechanisms:
- In E. coli and K. pneumoniae, enhanced binding to penicillin-binding proteins (PBPs) emerges as a critical factor contributing to improved bacterial inhibition [45].
- Against P. aeruginosa and A. baumannii, increased uptake appears to be the primary determinant of enhanced activity [45].
- SACs demonstrate reduced susceptibility to certain β-lactamases and efflux pumps, though the extent of this protection varies by bacterial species and enzyme class [45].
Table: Impact of Iron Concentration on Sideromycin Activity (MIC, μM)
| Bacterial Strain | Conjugate | Standard Medium | Iron-Depleted Medium | Iron-Supplemented Medium |
|---|---|---|---|---|
| E. coli | BAMP | 0.2 | 0.1 | 25 |
| K. pneumoniae | BLOR | 0.4 | 0.2 | >100 |
| P. aeruginosa | BAMP | 1.6 | 0.1 | 50 |
| A. baumannii | MCEF | 1.6 | 0.2 | >100 |
The profound impact of iron availability on SAC efficacy underscores the importance of the host-pathogen iron competition dynamic. In iron-depleted environments that mimic host conditions, SACs achieve their maximal antibacterial activity due to upregulated TBDT expression [45].
Experimental Approaches and Methodologies
Standardized Assessment Protocols
Determination of Minimum Inhibitory Concentrations (MICs) under Iron-Modified Conditions
- Prepare cation-adjusted Mueller-Hinton broth treated with Chelex 100 to create iron-depleted medium [45]
- Validate iron depletion using growth curves with incremental iron supplementation (0, 0.1, 1.0 μg/mL FeCl₃) [45]
- Perform broth microdilution MIC assays according to CLSI guidelines alongside iron-replete controls
- Include relevant reference strains and quality control organisms in each assay
Evaluation of Outer Membrane Permeation
- Employ engineered strains with defined porin deficiencies and TBDT deletions
- Utilize fluorescently labeled siderophore analogs to quantify uptake kinetics via flow cytometry
- Conduct competition assays with native siderophores to assess receptor specificity
- Implement radiolabeled antibiotic warheads to directly measure intracellular accumulation
Analysis of Target Engagement
- Express and purify recombinant PBPs for surface plasmon resonance binding studies
- Perform Bocillin FL competition assays to quantify PBP binding affinity
- Utilize β-lactamase hydrolysis assays to assess stability to enzymatic inactivation
- Employ efflux pump knockout strains to delineate transport versus retention contributions
Advanced Mechanistic Studies
Structural Characterization of Receptor-Ligand Interactions
- Purify TBDTs in native conformation using detergent solubilization protocols
- Conduct X-ray crystallography of TBDT-SAC complexes to determine binding modes
- Employ cryo-EM for structural analysis of larger transport complexes
- Utilize molecular dynamics simulations to model transport mechanisms
In Vivo Efficacy Assessment
- Utilize murine thigh infection models with iron dextran pretreatment to modulate host iron status
- Implement neutropenic animal models for disseminated infection studies
- Measure bacterial burden reduction in target tissues after SAC administration
- Determine pharmacokinetic/pharmacodynamic indices to guide dosing strategies
The Scientist's Toolkit: Essential Research Reagents
Table: Key Reagents for Sideromycin Research
| Reagent Category | Specific Examples | Research Application | Technical Considerations |
|---|---|---|---|
| Bacterial Strains | E. coli BW25113 (WT and isogenic TBDT mutants); P. aeruginosa PAO1; A. baumannii ATCC 19606; ESKAPE pathogen panel | Mechanism of action studies; spectrum of activity assessment | Include clinical MDR isolates; verify iron-responsive gene expression |
| Growth Media | Chelex 100-treated MHB; iron-depleted defined minimal media; iron supplement solutions (FeCl₃, FeSO₄) | Iron restriction studies; induction of siderophore uptake systems | Validate iron depletion via growth kinetics; monitor for contamination |
| Reference Compounds | Natural siderophores (enterobactin, ferrichrome); synthetic SACs (cefiderocol, BAMP, BLOR); unconjugated antibiotics | Uptake competition assays; structure-activity relationship studies | Verify purity and stability; prepare fresh stock solutions |
| Analytical Standards | Deuterated siderophore analogs; fluorescent siderophore conjugates (BODIPY-fl-catechol) | Quantification of uptake kinetics; receptor binding studies | Optimize detection methods (HPLC-MS, fluorescence polarization) |
| Enzymatic Tools | Recombinant PBPs (PBP1a, PBP2, PBP3); β-lactamases (NDM-1, KPC-2, OXA-48); periplasmic peptidases | Target engagement studies; stability assessment; linker cleavage evaluation | Maintain protein activity with proper storage conditions |
Future Directions and Clinical Translation
The field of siderophore-mediated drug delivery continues to evolve with several promising research frontiers:
Expanded Warhead Diversity: While β-lactams have dominated SAC development, recent work explores conjugation with fluoroquinolones, aminoglycosides, and novel antibacterial agents targeting intracellular processes [50] [49].
Narrow-Spectrum Approaches: Engineering SACs with specificity for particular bacterial pathogens by mimicking their unique endogenous siderophores offers potential for microbiome-sparing therapy [45] [48].
Nanotechnology Integration: Combining SAC strategies with nanoparticle delivery systems may further enhance targeting specificity and therapeutic index while reducing potential off-target effects [47] [52].
Diagnostic Applications: Siderophore-fluorophore conjugates show promise as imaging agents for specific detection of bacterial infections, potentially enabling rapid pathogen identification and treatment monitoring [48].
Despite the clinical success of cefiderocol, challenges remain in SAC development, including the potential for resistance through TBDT mutations, optimization of linker chemistry for specific bacterial enzyme cleavage, and balancing broad-spectrum activity with target selectivity [50] [49]. Continued research into the structural basis of siderophore recognition and transport will guide the rational design of next-generation conjugates with enhanced efficacy against the most recalcitrant multidrug-resistant pathogens.
Siderophore mimicry and Trojan horse strategies represent a paradigm shift in overcoming the permeability barrier of Gram-negative bacteria. By hijacking essential bacterial iron acquisition systems, these approaches transform a fundamental host-pathogen interaction into an effective drug delivery platform. The structural insights gleaned from natural sideromycins, combined with advanced synthetic chemistry and mechanistic understanding of transport systems, have yielded conjugates with remarkable potency against multidrug-resistant pathogens. As outer membrane permeability continues to pose a fundamental challenge in antibiotic discovery, leveraging active iron transport mechanisms through siderophore-antibiotic conjugates offers a promising pathway to address the escalating crisis of antimicrobial resistance.
The outer membrane (OM) of Gram-negative bacteria constitutes a formidable barrier to antimicrobial agents, significantly contributing to intrinsic antibiotic resistance. A key mechanism for overcoming this barrier is the "self-promoted uptake" pathway, utilized primarily by cationic antimicrobial peptides such as polymyxins. This pathway involves the displacement of divalent cations that stabilize the lipopolysaccharide (LPS) matrix, leading to increased OM permeability. This technical guide explores the molecular basis of self-promoted uptake, detailing the structural interactions, experimental methodologies for investigation, and its implications for antibiotic resistance research. Framed within the broader context of outer membrane permeability, this analysis provides researchers with a comprehensive resource on a critical mechanism for penetrating the Gram-negative bacterial envelope.
The outer membrane of Gram-negative bacteria is an asymmetric bilayer that provides exceptional protection against external threats. Its inner leaflet consists of phospholipids, while the outer leaflet is composed primarily of lipopolysaccharides (LPS) [10] [26]. This unique architecture is a major determinant of the inherent resistance of Gram-negative species to many antibiotics.
LPS molecules are complex glycolipids consisting of three domains: the hydrophobic lipid A anchor, a core oligosaccharide, and a distal O-antigen polysaccharide chain [10]. The dense packing of LPS molecules with their saturated fatty acid chains creates a low-fluidity barrier that effectively excludes hydrophobic compounds [26]. Furthermore, the anionic groups present on the LPS core and lipid A components are cross-bridged by divalent cations (Mg²⁺ and Ca²⁺), which neutralize electrostatic repulsion between adjacent LPS molecules and stabilize the membrane structure [10]. This organization creates a formidable obstacle that antibiotics must overcome to reach their intracellular targets.
Molecular Mechanism of Self-Promoted Uptake
The Cationic Displacement Model
Self-promoted uptake represents a sophisticated countermeasure to LPS barrier function. Cationic antimicrobial peptides (CAPs), such as polymyxin B and colistin (polymyxin E), exploit the electrostatic properties of the OM to facilitate their entry. These molecules are characterized by a polycationic nature and contain both hydrophilic and hydrophobic domains [53] [54].
The mechanism proceeds through sequential steps:
Electrostatic Attraction: The positively charged groups on CAPs are attracted to the negatively charged phosphate and carboxyl groups on lipid A and the core oligosaccharide of LPS [10] [54].
Competitive Cation Displacement: The CAPs competitively displace the divalent cations (Mg²⁺ and Ca²⁺) that bridge adjacent LPS molecules [10]. This displacement occurs because the affinity of the cationic peptides for the anionic sites exceeds that of the divalent cations.
Membrane Destabilization: The removal of stabilizing cations increases the lateral repulsion between LPS molecules, disrupting the tight packing of the outer leaflet and creating localized disruptions in membrane integrity [10].
Cellular Uptake: The peptide molecules then insert into the membrane, either traversing it to reach the periplasm or inner membrane, or creating temporary portals that facilitate the uptake of other antimicrobial agents [10] [53].
Table 1: Key Characteristics of Cationic Antibiotics Utilizing Self-Promoted Uptake
| Antibiotic | Net Charge | Primary LPS Target | Clinical Significance |
|---|---|---|---|
| Polymyxin B | +5 | Lipid A phosphates | Last-resort antibiotic |
| Colistin (Polymyxin E) | +5 | Lipid A phosphates | Last-resort antibiotic |
| Polymyxin B Nonapeptide (PMBN) | +5 | Lipid A phosphates | Research tool (permeabilizer) |
| Designed Cationic Peptides | Variable (+4 to +7) | Core oligosaccharide/Lipid A | Experimental therapeutics |
Structural Determinants of LPS-Antibiotic Interaction
The efficiency of self-promoted uptake depends critically on the molecular features of both the antibiotic and LPS:
Charge Distribution: Recent studies with designed cationic peptides derived from Ponericin W1 demonstrate that clustering cationic residues (e.g., lysine) at the N- or C-terminus, creating "linear amphipathicity," enhances binding to LPS compared to interspersing charged residues throughout the sequence [54]. This clustered arrangement may mimic the natural organization of polymyxins.
Hydrophobicity Balance: While cationic charge facilitates initial binding, sufficient hydrophobicity is required for subsequent membrane insertion. However, excessive hydrophobicity can increase toxicity to host cells, necessitating a careful balance [54].
LPS Structural Variations: Bacteria can modify their LPS structure to reduce its net negative charge, thereby decreasing affinity for cationic antibiotics. Common modifications include the addition of 4-amino-4-deoxy-L-arabinose (L-Ara4N) or phosphoethanolamine to lipid A phosphate groups, which are regulated by the PmrA-PmrB two-component system in Salmonella and E. coli [10].
Experimental Analysis of Self-Promoted Uptake
Key Methodologies and Protocols
Investigating self-promoted uptake requires multidisciplinary approaches that assess both binding interactions and functional consequences:
1. LPS Binding Assays
- Objective: Quantify antibiotic binding to isolated LPS or whole cells.
- Protocol: Use fluorescently tagged antibiotics (e.g., dansyl-polymyxin) or radiolabeled compounds. Incubate with LPS or bacterial cells, separate bound from unbound antibiotic via centrifugation or filtration, and measure bound fraction. Polymyxin-resistant mutants have been shown to bind only 25% of the antibiotic compared to wild-type strains [10].
- Applications: Determine binding constants, competitive inhibition studies with divalent cations.
2. Outer Membrane Permeabilization Assays
- Objective: Evaluate functional disruption of OM integrity.
- Protocol: Measure increased sensitivity to hydrophobic antibiotics (e.g., novobiocin, fusidic acid) or uptake of hydrophobic fluorescent dyes (e.g., N-phenyl-1-naphthylamine). Treatment with permeabilizers like polymyxin B nonapeptide (PMBN) can increase sensitivity to hydrophobic antibiotics by tens- to hundreds-fold [10].
- Applications: Assess structure-activity relationships of cationic peptides, study bacterial resistance mechanisms.
3. Liposome Leakage Studies
- Objective: Model membrane disruption in a controlled system.
- Protocol: Prepare liposomes incorporating purified LPS in their membrane, encapsulate a fluorescent dye (e.g., calcein), and monitor dye release upon antibiotic addition.
- Applications: Elucidate mechanisms of membrane disruption without complications from cellular processes.
4. Minimum Inhibitory Concentration (MIC) Determinations
- Objective: Evaluate antibacterial potency under standardized conditions.
- Protocol: Broth microdilution according to CLSI guidelines. Test against clinical isolates, including multidrug-resistant strains. For P. aeruginosa, cationic peptides with clustered lysine residues show MIC values as low as 1 μM [54].
- Applications: Correlate structural features with biological activity, screen novel compounds.
Table 2: Quantitative Measurements of Membrane Permeabilization Effects
| Permeabilizing Agent | Target Bacteria | Effect on Hydrophobic Antibiotic Sensitivity | Magnitude of Effect |
|---|---|---|---|
| Polymyxin B Nonapeptide (PMBN) | E. coli, S. typhimurium | Novobiocin, Fusidic acid, Clindamycin | 10 to 100-fold increase in sensitivity [10] |
| Tris/EDTA | E. coli, S. typhimurium | Various hydrophobic antibiotics | Similar to deep rough mutants [10] |
| "Deep Rough" LPS mutants | E. coli, S. typhimurium | Hydrophobic antibiotics, detergents, bile salts | High intrinsic sensitivity [10] |
| Cationic Peptides (PonN series) | MDR P. aeruginosa | Intrinsic activity | MIC ~1-4 μM [54] |
Visualizing the Mechanism: Molecular Pathways
The following diagram illustrates the sequential process of self-promoted uptake, from initial binding to membrane disruption:
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Studying Self-Promoted Uptake
| Reagent/Category | Specific Examples | Research Application |
|---|---|---|
| Cationic Antibiotics | Polymyxin B, Colistin, Polymyxin B nonapeptide (PMBN) | Positive controls, mechanism studies [10] |
| Engineered Peptides | Ponericin W1 derivatives (PonN, PonC, PonAmp) | Structure-activity relationship studies [54] |
| Membrane Probes | N-phenyl-1-naphthylamine (NPN), 1-N-phenylnaphthylamine | Measure outer membrane permeabilization [10] |
| LPS Modifiers | EDTA, Tris-EDTA | Control for LPS disruption, cation chelation [10] |
| Bacterial Strains | Deep rough mutants (Ra to Re chemotypes), PmrA/PmrB mutants | Study LPS structure-function relationships [10] |
| Ionophores | Valinomycin (K⁺), CCCP (H⁺) | Study ion effects on membrane permeability [24] |
Bacterial Resistance Mechanisms
Bacteria have evolved sophisticated countermeasures to subvert self-promoted uptake, primarily through modifications to the LPS target:
LPS Remodeling: The most common resistance mechanism involves the addition of cationic groups to lipid A phosphates. The PmrA-PmrB regulated addition of 4-amino-4-deoxy-L-arabinose (L-Ara4N) and/or phosphoethanolamine neutralizes the negative charges, reducing the initial electrostatic attraction for cationic antibiotics [10]. These modifications can reduce polymyxin binding by up to 75% in resistant mutants [10].
Regulatory Systems: Environmental sensing through two-component systems (e.g., PmrA-PmrB, PhoP-PhoQ) allows bacteria to dynamically modify their LPS in response to threatening conditions, including the presence of cationic antimicrobial peptides [10].
Outer Membrane Vesicles (OMVs): Bacteria can actively release OM fragments containing LPS and bound antibiotics through vesiculation, effectively reducing the local concentration of antimicrobial agents [26].
Research Implications and Future Directions
Understanding self-promoted uptake at a molecular level opens several promising research avenues:
Novel Antibiotic Design: The principles of self-promoted uptake can inform the development of new cationic peptides with optimized charge distribution and hydrophobicity. The demonstrated enhanced activity of lysine-clusted peptides against multidrug-resistant P. aeruginosa provides a template for such design efforts [54].
Combination Therapies: Permeabilizing agents like PMBN that employ self-promoted uptake can be paired with conventional antibiotics that otherwise struggle to penetrate the OM, creating synergistic combinations that overcome permeability barriers [10].
Resistance Breakers: Inhibitors of the bacterial enzymes responsible for LPS modification (e.g., ArnT for L-Ara4N transfer) could restore sensitivity to existing cationic antibiotics, representing an attractive adjuvant strategy.
Delivery Platforms: The OM permeabilization capability of cationic peptides can be harnessed to improve the delivery of other therapeutic agents, including traditional antibiotics and nucleic acids.
Self-promoted uptake represents a critical paradigm in the interplay between Gram-negative bacteria and antimicrobial agents. By targeting the fundamental ionic interactions that stabilize the outer membrane, cationic antibiotics create a foothold for their own penetration and potentially for other therapeutics. While bacterial resistance through LPS modification presents a significant clinical challenge, detailed understanding of this pathway continues to inspire innovative strategies to circumvent the formidable barrier of the Gram-negative outer membrane. As research advances, the principles of self-promoted uptake will undoubtedly remain central to efforts in overcoming antibiotic resistance in Gram-negative pathogens.
The outer membrane of Gram-negative bacteria presents a formidable barrier to antibiotic penetration, contributing significantly to the growing crisis of antimicrobial resistance. Porins, the water-filled β-barrel proteins in the outer membrane, have long been recognized as the primary gateways for hydrophilic antibiotics to reach their intracellular targets. However, bacterial pathogens have evolved sophisticated mechanisms to reduce porin expression or alter porin function, effectively limiting antibiotic entry and conferring resistance. Consequently, the exploration of non-classical uptake pathways represents a paradigm shift in overcoming antibiotic resistance. This whitepaper synthesizes recent advances in understanding and exploiting alternative penetration mechanisms, including nutrient transporter systems and completely porin-independent routes. These innovative strategies promise to circumvent traditional resistance mechanisms and restore the efficacy of existing antibiotics against multidrug-resistant pathogens.
The imperative for these approaches is underscored by the critical status of carbapenem-resistant Enterobacteriaceae and Pseudomonas aeruginosa, classified by the World Health Organization as priority pathogens of global concern [55]. As conventional porin-mediated pathways become compromised through bacterial adaptation, the field must increasingly focus on unconventional uptake mechanisms that bacteria cannot as easily regulate. This review examines three promising non-classical pathways: (1) outer membrane vesicle-mediated antibiotic delivery, (2) metalloantibiotic exploitation of direct membrane diffusion, and (3) metabolic modulation of membrane permeability. For each pathway, we provide quantitative analyses of transport efficiency, detailed experimental protocols for investigation, and essential research tools to advance this emerging field.
Outer Membrane Vesicle-Mediated Antibiotic Delivery
Mechanism and Experimental Evidence
Outer membrane vesicles (OMVs) are nanosized, spherical structures naturally released by Gram-negative bacteria during their growth. These vesicles form through bulging of the outer membrane, encapsulating periplasmic content before pinching off from the bacterial surface. Recent research has demonstrated that OMVs can function as efficient delivery vehicles for antibiotic transport, completely bypassing the need for porin-mediated passage [56].
In a groundbreaking study, Wu et al. investigated the ability of imipenem-encapsulated OMVs to inhibit the growth of multidrug-resistant Gram-negative clinical isolates [56]. The researchers isolated OMVs from E. coli cultures and employed electroporation to load the vesicles with imipenem. Remarkably, OMV-encapsulated imipenem exhibited significantly enhanced antibacterial activity compared to free antibiotic, with the minimum inhibitory concentration (MIC) reduced by up to 8-fold against porin-deficient strains. Crucially, this delivery mechanism remained effective even in a panel of porin knockout strains, confirming its porin-independent nature. This finding suggests that OMVs likely fuse with the bacterial outer membrane or are internalized through endocytosis-like processes, directly depositing their cargo into the periplasmic space without requiring passage through porin channels.
Table 1: Efficacy of OMV-Encapsulated Imipenem Against Porin-Deficient Strains
| Bacterial Strain | Porin Status | Free Imipenem MIC (μg/mL) | OMV-Imipenem MIC (μg/mL) | Fold Improvement |
|---|---|---|---|---|
| E. coli WT | Wild-type | 0.5 | 0.25 | 2x |
| E. coli ΔompC | OmpC deficient | 2.0 | 0.25 | 8x |
| E. coli ΔompF | OmpF deficient | 4.0 | 0.5 | 8x |
| E. coli ΔompC/ΔompF | Double knockout | 16.0 | 2.0 | 8x |
| K. pneumoniae MDR | Clinical isolate | 32.0 | 4.0 | 8x |
Protocol: OMV Isolation and Antibiotic Loading
OMV Isolation Protocol:
- Grow E. coli in appropriate medium (e.g., LB broth) to late exponential phase (OD600 ≈ 0.8).
- Separate cells from culture supernatant by centrifugation at 10,000 × g for 20 minutes at 4°C.
- Filter the supernatant through a 0.45 μm pore-size membrane to remove residual cells.
- Concentrate the vesicles from the filtered supernatant using tangential flow filtration with a 100 kDa molecular weight cut-off membrane.
- Purify OMVs by ultracentrifugation at 150,000 × g for 2 hours at 4°C.
- Resuspend the OMV pellet in sterile phosphate-buffered saline (PBS) and store at 4°C for immediate use or -80°C for long-term storage.
Antibiotic Loading via Electroporation:
- Mix 500 μL of OMV suspension (approximately 1-5 mg/mL protein content) with 50 μg of imipenem.
- Transfer the mixture to a pre-chilled 2 mm electroporation cuvette.
- Perform electroporation at 2.5 kV, 25 μF, and 200 Ω.
- Immediately transfer the electroporated sample to ice for 10 minutes.
- Remove non-encapsulated antibiotic by gel filtration using a Sephadex G-50 column.
- Verify antibiotic encapsulation efficiency via high-performance liquid chromatography (HPLC).
Metalloantibiotics: Porin-Independent Membrane Diffusion
Mechanism and Permeation Advantages
Metalloantibiotics represent an innovative class of antimicrobial agents that bypass porin-dependent uptake pathways through enhanced membrane permeability. These compounds are ternary complexes formed between fluoroquinolone antibiotics (such as ciprofloxacin), copper ions, and phenanthroline auxiliary ligands [57]. The resulting complexes exhibit distinct physicochemical properties that favor direct diffusion across the lipid bilayer of the bacterial outer membrane.
Biophysical studies comparing ciprofloxacin and its copper complex (CuCpxPhen) revealed striking differences in their permeation mechanisms [57]. While ciprofloxacin primarily relies on OmpF porins for outer membrane translocation, the metalloantibiotic demonstrates significantly reduced interaction with this major porin. X-ray crystallography of OmpF porin crystals soaked with CuCpxPhen showed no well-defined binding site for the complex, indicating weak porin interaction. Molecular dynamics simulations further demonstrated that the metalloantibiotic faces a much higher free energy barrier (approximately 3-fold greater) when crossing the constriction zone of OmpF compared to unmodified ciprofloxacin. This unfavorable interaction energetically drives the metalloantibiotic toward the porin-independent lipid-mediated uptake pathway.
Table 2: Permeation Properties of Fluoroquinolones vs. Metalloantibiotics
| Parameter | Ciprofloxacin | CuCpxPhen (Metalloantibiotic) |
|---|---|---|
| Primary uptake route | Porin-mediated (OmpF) | Lipid bilayer diffusion |
| Free energy barrier through OmpF | Low | High (≈3x greater) |
| Permeability in porin-mimetic system | High | Low |
| Membrane partition coefficient | Low | High (≈5x greater) |
| Efficacy in porin-deficient strains | Reduced | Maintained |
| Minimum inhibitory concentration (MIC) in OmpF-deficient E. coli | Increased 8-fold | Unchanged |
Protocol: Assessing Porin-Independent Uptake
Metalloantibiotic Synthesis:
- Dissolve ciprofloxacin (1 mmol) in methanol:water (1:1 v/v) with slight heating if necessary.
- Add 1.1 mmol of 1,10-phenanthroline with stirring.
- Slowly add 1 mmol of copper(II) chloride while maintaining constant stirring.
- Adjust pH to 7.0-7.5 with dilute NaOH and continue stirring for 4 hours at room temperature.
- Recover the precipitate by filtration and wash with cold methanol.
- Characterize the complex using UV-Vis spectroscopy, mass spectrometry, and elemental analysis.
Porin-Mimetic Permeability Assay:
- Prepare a phospholipid bilayer using a commercial system or by creating liposomes from E. coli lipid extract.
- Incorporate OmpF porin into the bilayer following established protocols for membrane protein reconstitution.
- Measure the flux of antibiotics and metalloantibiotics across the membrane using a diffusion cell apparatus.
- Sample the receiver compartment at regular intervals and quantify compound concentration using HPLC.
- Calculate permeability coefficients from the flux data using Fick's law of diffusion.
Molecular Dynamics Simulations:
- Obtain the crystal structure of OmpF porin from the Protein Data Bank (PDB ID: 2OMF).
- Parameterize the metalloantibiotic using appropriate force fields (e.g., GAFF for organic components).
- Embed the porin in a realistic lipid bilayer (e.g., POPE/POPG mixture).
- Solvate the system with water molecules and add ions to physiological concentration.
- Perform umbrella sampling simulations along the porin channel axis to calculate potential of mean force.
- Analyze trajectories to determine free energy barriers and preferred pathways.
Metabolic Modulation of Membrane Permeability
Ionic Regulation of Porin Function
Beyond completely porin-independent pathways, research has revealed that bacteria dynamically regulate porin permeability through metabolic activity, presenting opportunities for intervention. Caño Muñiz et al. demonstrated that porin permeability in Escherichia coli is controlled by changes in periplasmic H+ and K+ concentrations influenced by metabolic state [24]. This ionic regulation occurs through charge-based effects on the porin pore diameter, providing a mechanism for bacteria to balance nutrient uptake with energy conservation.
Using single-cell imaging with genetically encoded ion sensors, the researchers observed that conductance through porins increases during starvation (low periplasmic H+), promoting nutrient uptake, and decreases during growth on lipid media (periplasmic acidification), limiting proton loss [24]. Importantly, high metabolic activity during growth in glucose media activates the inner membrane voltage-gated potassium channel Kch, increasing periplasmic potassium and enhancing porin permeability. This metabolic control explains observed increases in ciprofloxacin resistance in bacteria catabolizing lipids and identifies Kch as a potential therapeutic target to improve bacterial killing by antibiotics. Molecular dynamics simulations suggested that protonation of amino acid residues on the periplasmic surface of OmpC reduces its pore diameter, providing a structural basis for this regulation.
Metabolic State-Driven Sensitization
The metabolic state-driven approach to combat antibiotic resistance exploits the fact that exogenous nutrient metabolites can stimulate antibiotic uptake through metabolic reprogramming [58]. This approach identifies crucial metabolic differences between antibiotic-sensitive and antibiotic-resistant bacteria, then uses specific nutrients to reprogram resistant cells to a sensitive state.
Peng et al. demonstrated that kanamycin-resistant Edwardsiella tarda shows low intracellular glucose and alanine levels compared to sensitive strains [58]. Exposure to exogenous alanine, glucose, or fructose restored susceptibility to kanamycin by activating the pyruvate cycle, increasing NADH production, proton motive force (PMF), and drug uptake. Similarly, Zhang et al. showed that exogenous glucose reprograms gentamicin-resistant Vibrio alginolyticus by stimulating the pyruvate cycle, NADH, PMF, ROS production, and gentamicin uptake [58]. These findings suggest that metabolic modulation can enhance antibiotic penetration regardless of porin status, potentially by increasing membrane potential or activating alternative uptake systems.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Studying Non-Classical Uptake Pathways
| Reagent | Function/Application | Example Use |
|---|---|---|
| 2NBDG (2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose) | Fluorescent glucose analog for tracking porin permeability [24] | Measure real-time changes in porin activity in response to metabolic shifts |
| Bocillin FL | Fluorescent penicillin analog for β-lactam penetration studies [24] | Visualize antibiotic penetration through different pathways |
| pHuji and pHluorin | Genetically encoded pH sensors for periplasmic and cytoplasmic pH monitoring [24] | Track periplasmic acidification and its effect on porin function |
| GINKO1 and GINKO2 | Genetically encoded potassium sensors [24] | Monitor potassium fluctuations in cytoplasm and periplasm |
| QuasAr2 | Genetically encoded voltage sensor [24] | Measure inner membrane potential changes |
| ArchT | Light-activated proton pump [24] | Optogenetically control periplasmic pH to manipulate porin permeability |
| Valinomycin | Potassium ionophore [24] | Artificially alter periplasmic K+ concentrations |
| CCCP (Carbonyl cyanide m-chlorophenyl hydrazone) | Protonophore [24] | Dissipate proton motive force to study its role in uptake |
| KEIO collection | E. coli single-gene knockout mutants [24] | Screen for genes involved in porin regulation and alternative uptake |
Conceptual Framework and Experimental Pathways
The diagram below illustrates the logical relationships between the non-classical uptake strategies, their mechanisms, and appropriate experimental validation approaches discussed in this review.
Non-Classical Antibiotic Uptake Pathways
The exploration of non-classical uptake pathways represents a transformative approach to overcoming antibiotic resistance in Gram-negative bacteria. The strategies discussed—outer membrane vesicle-mediated delivery, metalloantibiotic development, and metabolic modulation of membrane permeability—each offer distinct advantages for bypassing porin-based resistance mechanisms. As research advances, the integration of these approaches with conventional antibiotic development promises to open new frontiers in antimicrobial therapy. In particular, the targeted manipulation of bacterial metabolic networks to enhance antibiotic uptake presents an opportunity to develop adjuvant therapies that restore susceptibility to existing antibiotics. Similarly, the continued refinement of vesicle-based delivery systems and metalloantibiotic chemistry may yield platforms adaptable to multiple antibiotic classes. Future work should focus on optimizing these strategies for clinical application, particularly in addressing the challenge of bacterial heterogeneity and evolutionary counter-adaptations.
The escalating crisis of antimicrobial resistance (AMR), particularly among Gram-negative pathogens, underscores the critical need for innovative therapeutic strategies. A significant barrier to effective treatment is the formidable outer membrane (OM), which restricts antibiotic accumulation within bacterial cells. This whitepaper delineates the role of OM-disrupting permeability enhancers in overcoming intrinsic, acquired, and spontaneous antibiotic resistance. We detail the mechanisms of various permeabilizing agents—including peptides, chelators, and small molecules—and their synergistic action with conventional antibiotics. Supported by recent experimental data and mechanistic studies, this review provides a framework for the rational design of combination therapies, offering a promising pathway to revitalize existing antibiotics and combat multidrug-resistant infections.
Antimicrobial resistance poses a catastrophic global threat, with drug-resistant infections causing an estimated 4.95 million deaths annually and projections of up to 10 million deaths per year by 2050 [32]. Gram-negative bacteria—such as Pseudomonas aeruginosa, Acinetobacter baumannii, and carbapenem-resistant Enterobacterales—present a particularly formidable challenge due to their complex cell envelope architecture [26]. The outer membrane (OM) of these pathogens functions as a sophisticated, asymmetric bilayer that constitutes a primary line of defense against antimicrobial agents [10].
The OM's outer leaflet is composed primarily of lipopolysaccharides (LPS), which are stabilized by divalent cations (Mg²⁺ and Ca²⁺) that bridge adjacent LPS molecules. This structure creates a densely packed, rigid barrier with a strong negative charge that effectively excludes many hydrophobic and large hydrophilic compounds [10] [26]. The inner leaflet contains phospholipids, and the membrane is punctuated by protein channels known as porins that permit selective passage of small, hydrophilic nutrients. This unique organization establishes two major permeation pathways: a lipid-mediated route for hydrophobic molecules, and porin-mediated diffusion for hydrophilic compounds [10]. The synergy between the low-permeability OM and broadly-specific efflux pumps creates a highly effective barrier-intruder clearance system, reducing intracellular antibiotic accumulation by several orders of magnitude [59]. Consequently, overcoming the OM permeability barrier represents a paramount challenge in antibiotic discovery and development, with permeability enhancers emerging as a pivotal strategy to potentiate existing antibiotics and extend their therapeutic utility against resistant pathogens.
Mechanisms of Outer Membrane Disruption
Outer membrane disruptors employ diverse molecular strategies to compromise membrane integrity, thereby facilitating antibiotic entry. These mechanisms can be broadly categorized as follows:
Cationic Peptide-Mediated Disruption
Cationic antimicrobial peptides (AMPs), including colistin and its derivative SPR741, as well as scorpion-derived peptides like AaeAP2a, utilize a "self-promoted uptake" mechanism [59]. These molecules are attracted to the negatively charged LPS via electrostatic interactions. They competitively displace the divalent cations that stabilize LPS-LPS interactions, disrupting membrane integrity and creating transient openings through which other antibiotics can pass [60] [61]. Colistin, a last-resort polymyxin antibiotic, exerts its activity through its polycationic peptide structure, which displaces cationic bridges between LPS molecules, followed by insertion of its hydrophobic tail into the membrane [32]. The novel peptide AaeAP2a has been shown to disrupt both inner and outer bacterial membranes, increasing membrane permeability and triggering metabolic collapse [61].
Chelator-Mediated Disruption
Chelating agents such as ethylenediaminetetraacetic acid (EDTA) function by sequestering the divalent cations (Mg²⁺ and Ca²⁺) that cross-bridge and stabilize adjacent LPS molecules [32] [10]. By removing these essential cations, EDTA weakens the cohesive forces between LPS molecules, leading to destabilization of the OM bilayer, increased membrane fluidity, and the formation of patches that are more permeable to hydrophobic compounds [10] [60]. This chelation effect can result in massive release of LPS into the surrounding medium [10].
Small Molecule Disruptors
Small organic molecules like pentamidine and polyaminoisoprenyl derivatives (e.g., NV716) represent another class of OM perturbants. NV716 binds directly to LPS and induces OM destabilization [32], while pentamidine, like cationic peptides, disrupts the cation bridging between LPS molecules [60]. These compounds typically exhibit sub-MIC activity when used as potentiators, meaning they permeabilize the OM without exhibiting significant bactericidal activity on their own.
The following table summarizes the primary classes of OM disruptors and their characteristics:
Table 1: Major Classes of Outer Membrane Disruptors
| Class | Representative Agents | Mechanism of Action | Key Features |
|---|---|---|---|
| Cationic Peptides | Colistin, SPR741, AaeAP2a, Squalamine | Displace divalent cations bridging LPS; integrate into membrane via electrostatic and hydrophobic interactions | "Self-promoted uptake" pathway; often derived from natural sources (e.g., scorpion venom) [32] [61] |
| Chelators | EDTA (Ethylenediaminetetraacetic acid) | Chelates Mg²⁺ and Ca²⁺ ions, destabilizing LPS lattice | Causes significant LPS release; well-established experimental tool [32] [10] |
| Small Molecules | NV716, Pentamidine | Bind directly to LPS, disrupting membrane organization and cation bridges | Synthetic compounds; can be optimized for pharmacological properties [32] [60] |
The molecular interplay between a permeation enhancer and the bacterial membrane is complex. The following diagram illustrates the generalized mechanism of outer membrane disruption, showing how different classes of agents overcome the LPS barrier to facilitate antibiotic entry:
Quantitative Assessment of Potentiation Efficacy
The potentiation capacity of OM disruptors is quantitatively evaluated by measuring the reduction in Minimum Inhibitory Concentration (MIC) of partner antibiotics. A ≥4-fold MIC reduction is typically considered significant potentiation [32] [60]. The efficacy of potentiation varies considerably based on the specific antibiotic-disruptor combination and the bacterial strain.
Recent studies have systematically screened multiple antibiotics in combination with different OM disruptors. For instance, against P. aeruginosa, the tetracycline doxycycline exhibited a 128-fold MIC reduction (from 64 mg/L to 0.5 mg/L) with the small molecule disruptor NV716, and a 64-fold reduction with EDTA [32]. Similarly, chloramphenicol (an amphenicol) showed a 16-fold MIC reduction with both NV716 and EDTA [32].
A broad screening study in E. coli demonstrated that OM perturbation potentiates a wide range of traditionally Gram-positive-active antibiotics, including macrolides (e.g., erythromycin, clarithromycin), rifamycins (e.g., rifampicin), and others like novobiocin, fusidic acid, and clindamycin [60]. The following table compiles quantitative potentiation data for various antibiotic classes:
Table 2: Efficacy of Outer Membrane Disruptors in Potentiating Antibiotic Activity
| Antibiotic Class | Example Agent | OM Disruptor | MIC Fold Reduction | Pathogen | Key Finding |
|---|---|---|---|---|---|
| Tetracyclines | Doxycycline | NV716 (10 µM) | 128-fold | P. aeruginosa | Strongest potentiation observed in this class [32] |
| Amphenicols | Chloramphenicol | EDTA (1 mM) | 16-fold | P. aeruginosa | Confirms OM permeability limits amphenicol activity [32] |
| Macrolides | Azithromycin | NV716 (10 µM) | 4-fold | P. aeruginosa | Variable potentiation for generally inactive macrolides [32] |
| Rifamycins | Rifampicin | SPR741 | >4-fold | E. coli | Consistently potentiated by all tested disruptors [60] |
| Glycopeptides | Vancomycin | EDTA | 32-fold | E. coli | Potentiation highly variable between disruptor types [60] |
| β-lactams | Meropenem | InC58 + AVI | 64-fold (MIC50 reduction) | Resistant Enterobacteria | Triple combination overcomes enzymatic resistance [62] |
Not all antibiotics benefit equally from OM disruption. Antibiotics that are intrinsically active against Gram-negative bacteria, such as fluoroquinolones and many β-lactams, typically show minimal potentiation, as their physicochemical properties already allow some degree of OM penetration, often through porin channels [60]. The most dramatic enhancements are observed for hydrophobic, large molecules (e.g., macrolides, rifamycins) whose entry is normally severely restricted by the intact OM barrier [60].
Experimental Methodologies for Evaluating OM Permeabilization
Minimum Inhibitory Concentration (MIC) Determination
The broth microdilution method, performed according to standards such as the CLSI M100-ED34, is the cornerstone for assessing potentiation [61].
Protocol:
- Inoculum Preparation: Isolate a single bacterial colony and inoculate it into Mueller-Hinton Broth (MHB). Incubate overnight at 37°C with agitation (e.g., 180 rpm).
- Standardization: Adjust the turbidity of the overnight culture to a 0.5 McFarland standard (approximately 1-2 x 10⁸ CFU/mL). Subsequently, dilute the suspension 1:100 in fresh MHB to achieve a working inoculum of ~1-2 x 10⁶ CFU/mL.
- Microdilution Plate Setup: In a 96-well plate, serially dilute the antibiotic (e.g., two-fold dilutions) in MHB across the rows. Include a sub-inhibitory concentration of the OM disruptor in all test wells. Controls must include (a) antibiotic alone (no disruptor), (b) disruptor alone (no antibiotic), and (c) growth control (media + inoculum, no agents).
- Inoculation and Incubation: Aliquot the standardized inoculum into each well (except sterility controls). Incubate the plate at 37°C for 16-20 hours.
- MIC Determination: The MIC is defined as the lowest concentration of antibiotic that completely prevents visible growth. The potentiation factor is calculated as MICantibiotic alone / MICantibiotic + disruptor [32] [61].
Time-Kill Kinetics Assay
This assay determines whether the combination of an OM disruptor and an antibiotic results in bactericidal (≥3-log10 CFU reduction) versus bacteriostatic activity.
Protocol:
- Culture Setup: Dilute a fresh bacterial culture in MHB and expose it to the antibiotic at multiples of the MIC (e.g., 0.5x, 1x, 2x, 4x MIC), both with and without a sub-MIC concentration of the OM disruptor. Include a control with no antimicrobial agents.
- Sampling and Enumeration: At predetermined time intervals (e.g., 0, 2, 4, 6, 8, 10, 12, 24 h), withdraw aliquots from each culture. Perform serial ten-fold dilutions in saline or PBS and plate onto Mueller-Hinton Agar (MHA).
- Incubation and Analysis: Incubate plates at 37°C for 24 hours, then enumerate the colonies. Plot log10 CFU/mL versus time to generate time-kill curves. Synergy is typically defined as a ≥2-log10 CFU/mL decrease with the combination compared to the most active single agent at 24 hours [61].
Membrane Integrity and Permeability Assays
SYTOX Green Uptake Assay: SYTOX Green is a fluorescent dye that is impermeant to intact bacterial membranes but readily enters cells with compromised membranes, binding to nucleic acids and exhibiting a significant fluorescence enhancement.
- Cell Preparation: Grow bacteria to mid-log phase, harvest by centrifugation, and wash/resuspend in an appropriate buffer (e.g., PBS or HEPES).
- Dye Addition: Incubate the bacterial suspension with SYTOX Green dye.
- Treatment and Measurement: Dispense the cell-dye mixture into a multi-well plate and treat with the OM disruptor. Monitor fluorescence intensity over time (e.g., every 5-10 minutes for 1-2 hours) using a microplate reader with excitation/emission wavelengths of ~504/523 nm. An increase in fluorescence indicates disruption of membrane integrity [61].
Other Mechanistic Probes:
- ATP Production Assays: Use luminometric assays to quantify intracellular ATP levels, as dissipation of membrane potential often collapses the proton motive force, reducing ATP synthesis.
- Reactive Oxygen Species (ROS) Detection: Employ fluorescent probes like H₂DCFDA or DHR123 to measure ROS accumulation, a common consequence of membrane stress and metabolic dysregulation.
- 1-N-phenylnaphthylamine (NPN) Uptake Assay: This hydrophobic fluorophore is excluded by the intact OM but enters upon disruption, fluorescing upon partitioning into the phospholipid inner membrane. Increased NPN fluorescence is a specific indicator of OM permeabilization.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Resources for OM Permeabilization Research
| Reagent / Resource | Function & Application | Examples & Key Details |
|---|---|---|
| OM Disruptors | Experimental agents to perturb outer membrane integrity | SPR741: Derivative of polymyxin B nonapeptide; completed Phase I trials [60]. NV716: Polyaminoisoprenyl small molecule; potent LPS binder [32]. AaeAP2a: Scorpion-derived antimicrobial peptide; disrupts both OM and IM [61]. |
| Reference Antibiotics | Partner drugs to assess potentiation efficacy | Doxycycline (Tetracycline): Shows high-fold MIC reduction [32]. Rifampicin (Rifamycin): Consistently potentiated across disruptors [60]. Novobiocin: Hydrophobic Gram-positive-active antibiotic [60]. |
| Fluorescent Probes | Detect membrane integrity and permeability changes | SYTOX Green: Impermeant nucleic acid stain for compromised membranes [61]. 1-N-phenylnaphthylamine (NPN): Hydrophobic probe for OM permeabilization. DCFDA / DHR123: Detect intracellular ROS accumulation [61]. |
| Cell Viability Assays | Quantify metabolic activity and cytotoxicity | WST-1 Assay: Measures cellular metabolic activity as a proxy for viability and potential cytotoxicity of enhancers [63]. Bacterial Colony Counting (CFU): Gold standard for determining bactericidal activity in time-kill assays [61]. |
| Strains & Culture Media | Provide biological context and growth environment | CRAB 236: Carbapenem-resistant A. baumannii clinical isolate from a One Health context (pet dog) [61]. Mueller-Hinton Broth/Agar (MHB/MHA): Standardized media for antimicrobial susceptibility testing [61]. |
The strategic disruption of the Gram-negative outer membrane represents a paradigm shift in combating antimicrobial resistance. By mitigating the primary permeability barrier, this approach rejuvenates the activity of existing antibiotics, particularly those limited by poor OM penetration. The compelling experimental evidence—demonstrating the ability of OM disruptors to overcome intrinsic, acquired, and spontaneous resistance—validates this strategy as a high-priority avenue for therapeutic development.
Future progress hinges on the rational design of optimized antibiotic-disruptor combinations based on a multidimensional understanding of physicochemical properties, including lipophilicity, molecular size, and polarity [32]. Furthermore, clinical translation requires rigorous assessment of resistance liability, toxicity, and pharmacokinetic/pharmacodynamic (PK/PD) compatibility in advanced infection models. As exemplified by the progression of SPR741 into clinical trials, the integration of OM permeabilizers into combination regimens holds immense potential to expand our therapeutic arsenal and confront the escalating threat of multidrug-resistant Gram-negative infections.
Resistance at the Gate: Diagnosing and Overcoming Permeability-Mediated Failure
The outer membrane of Gram-negative bacteria serves as a formidable barrier against antimicrobial agents, with porins acting as critical gatekeepers that govern the permeability of this protective layer. These β-barrel proteins facilitate the passive diffusion of essential nutrients, ions, and other small hydrophilic molecules while simultaneously creating an Achilles' heel through which antibiotics can enter the cell [64] [55]. In clinical settings, the strategic compromise of this entry route represents a fundamental resistance mechanism employed by major bacterial pathogens.
Porin deficiencies—encompassing downregulation, mutation, and complete loss-of-function—have emerged as significant contributors to multidrug resistance, particularly against last-resort antibiotics like carbapenems [64] [65]. The World Health Organization has recognized carbapenem-resistant Enterobacteriaceae and Pseudomonas aeruginosa as critical and high-priority pathogens, respectively, underscoring the urgent public health threat posed by these resistance mechanisms [64] [55]. This whitepaper examines the molecular basis, regulatory pathways, and clinical implications of porin deficiencies within the broader context of outer membrane permeability and antibiotic resistance research, providing technical guidance for scientists and drug development professionals confronting these challenges.
Porin Structure and Function
General Architecture and Physiological Roles
Porins are transmembrane proteins that form water-filled channels traversing the outer membrane of Gram-negative bacteria. Most general porins, such as OmpF and OmpC in Escherichia coli and their orthologs OmpK35 and OmpK36 in Klebsiella pneumoniae, adopt a characteristic homotrimeric configuration [64]. Each monomer consists of a 16-stranded β-barrel structure connected by extracellular loops and periplasmic turns [64] [66]. The third loop (L3) folds back into the channel, creating a constriction zone or eyelet that determines the size and charge selectivity of molecules that can pass through [64].
These proteins primarily facilitate the passive diffusion of hydrophilic nutrients, including sugars, ions, and amino acids, while simultaneously allowing waste products to exit the cell [64] [55]. Beyond their nutritional roles, porins contribute to membrane integrity through interactions with peptidoglycan, participate in bacterial adhesion and invasion, and help neutralize host defense mechanisms [64] [66]. The OmpA protein, for instance, interacts with peptidoglycan via its C-terminal domain, specifically through aspartate (D271) and arginine (R286) residues, thereby maintaining envelope stability [66].
Permeability Characteristics of Major Porins
Table 1: Properties and Functions of Major Porins in Gram-Negative Bacteria
| Porin | Structural Features | Primary Functions | Permeability Characteristics | Role in Resistance |
|---|---|---|---|---|
| OmpF/OmpK35 | 16-stranded β-barrel; larger pore constriction | Nutrient uptake; environmental sensing | More permeable; less cation-selective | Major entry route for β-lactams; often downregulated in resistance |
| OmpC/OmpK36 | 16-stranded β-barrel; slightly narrower pore | Osmoprotection; nutrient uptake in high osmolarity | Less permeable; more cation-selective | Reduced antibiotic entry; mutations alter selectivity |
| OmpA | Monomeric β-barrel with periplasmic domain | Membrane integrity; peptidoglycan binding | Non-specific slow porin | Reduces membrane permeability; contributes to intrinsic resistance |
| LamB | Substrate-specific porin; trimeric | Maltose and maltodextrin transport | Specific channel | Downregulation reduces antibiotic uptake; binds last-resort drugs |
The permeability differences between porin types significantly influence antibiotic penetration. OmpC is less permeable than OmpF due to both its slightly narrower pore and the presence of more negative charges in its pore lining region [64]. Research has demonstrated that 10 residues differing in charge between OmpF and OmpC account for their differential antibiotic permeation properties, with OmpC's increased cation selectivity explaining its lower permeability to anionic β-lactams like aztreonam and ceftriaxone [64].
Mechanisms of Porin Deficiency
Transcriptional and Post-transcriptional Regulation
Bacteria employ sophisticated regulatory systems to control porin expression in response to environmental cues. In Enterobacteriaceae, the EnvZ/OmpR two-component system serves as the primary regulator of OmpF and OmpC expression [64]. This system modulates porin production based on osmolarity through a phosphorylation-dependent mechanism:
- Under low-osmolarity conditions, minimal EnvZ phosphorylation occurs, resulting in low levels of phosphorylated OmpR (OmpR-P). This favors ompF transcription while ompC expression remains minimal [64].
- Under high-osmolarity conditions, EnvZ autophosphorylation increases, elevating OmpR-P levels. This activates ompC transcription while repressing ompF expression [64].
This reciprocal regulation provides an adaptive advantage, as the narrower OmpC pore offers enhanced protection against toxic molecules in hostile environments like the intestine, while OmpF facilitates efficient nutrient acquisition in nutrient-poor conditions [64].
Additional regulatory mechanisms further modulate porin expression. In Aeromonas veronii, the SmpB protein positively regulates OmpA expression during stationary phase by binding to specific regions of the OmpA promoter [66]. In Acinetobacter baumannii, the A1S_0316 protein acts as an anti-repressor by binding the OmpA promoter region with higher affinity than the global repressor H-NS [66]. Furthermore, the blue light-sensing protein BlsA influences OmpA expression under light conditions, affecting membrane permeability to lipophilic compounds [66].
Diagram 1: Regulatory pathway of porin expression through the EnvZ/OmpR two-component system in response to osmolarity changes.
Mutational Alterations and Functional Consequences
Porin function can be compromised through various mutational mechanisms that either reduce channel permeability or eliminate porin production entirely. These mutations arise under antibiotic selective pressure and represent a direct evolutionary response to antimicrobial challenge.
Structural mutations that modify the constriction zone without significantly altering pore size can dramatically impact antibiotic permeability. Research on clinical E. coli isolates revealed that sequential mutations in OmpC (OmpC20 to OmpC33 variants) resulted in modulated antibiotic transport without major changes to pore dimensions [67]. Molecular dynamics simulations indicated that these mutations perturb the transverse electrostatic field at the constriction zone, reducing the passage of antibiotics like cefotaxime through the pore [67]. This represents a sophisticated resistance mechanism that preserves nutrient uptake while limiting antibiotic penetration.
Loss-of-function mutations include frameshifts, nonsense mutations, and deletions that completely abolish porin production. In K. pneumoniae, clinical isolates frequently exhibit truncated OmpK35 and/or OmpK36 proteins due to mutations in their encoding genes [68] [69]. Whole-genome sequencing of 26 K. pneumoniae clinical isolates revealed that ompK35 was intact in only nine wild-type isolates, while it was truncated in 13 isolates and contained point mutations in others [69]. Similarly, ompK36 was truncated in two isolates and contained various mutations in the remainder [69]. This genetic heterogeneity highlights the diverse strategies bacteria employ to limit porin-mediated antibiotic entry.
Metabolic Control of Porin Permeability
Recent research has revealed that porin permeability is dynamically regulated by metabolic activity and ion concentrations. In E. coli, porin permeability is controlled by changes in periplasmic H+ and K+ concentrations mediated by metabolic processes [24].
- During starvation conditions, low periplasmic H+ increases porin conductance, promoting nutrient uptake [24].
- During growth on lipid media, periplasmic acidification decreases porin permeability, limiting proton loss and conserving energy [24].
- During growth on glucose media, high metabolic activity activates the voltage-gated potassium channel Kch, increasing periplasmic potassium and enhancing porin permeability to dissipate reactive oxygen species [24].
This metabolic regulation demonstrates how bacteria balance the competing demands of nutrient acquisition and energy conservation, with direct implications for antibiotic permeability. Single-cell imaging experiments using the fluorescent glucose analog 2NBDG revealed tight correlation between membrane voltage and porin permeability, with membrane depolarization resulting in increased porin-mediated transport [24].
Research Methodologies and Experimental Approaches
Techniques for Porin Expression Profiling
Accurate characterization of porin expression patterns is essential for understanding resistance mechanisms. Several methodological approaches enable comprehensive porin profiling:
SDS-PAGE analysis remains a fundamental technique for separating outer membrane proteins (OMPs) by molecular weight. Proteins are extracted using detergents like sodium lauroyl sarcosinate to solubilize inner membrane and non-integral outer membrane components, leaving integral OMPs for analysis [69]. Following separation, protein bands can be excised and identified by liquid chromatography coupled to mass spectrometry (LC-MS/MS) [69].
MALDI-TOF mass spectrometry offers a rapid alternative for OMP detection. A comparative study of 26 K. pneumoniae clinical isolates demonstrated that MALDI-TOF/MS reliably detected porins with results consistent with SDS-PAGE, showing peaks at ~35,700 (OmpA), ~37,000 (OmpK35), and ~38,000 (OmpK36) m/z [69]. This method is particularly valuable for clinical laboratories already equipped with MALDI-TOF systems for bacterial identification.
Whole-genome sequencing (WGS) enables comprehensive analysis of porin genes and identification of mutations. However, studies have shown that WGS cannot always anticipate protein expression due to complex post-transcriptional regulation [69]. While WGS reliably detects truncations and major mutations, it may miss regulatory changes affecting porin expression levels, highlighting the importance of complementary proteomic approaches.
Table 2: Comparison of Porin Profiling Methodologies
| Method | Principles | Applications | Limitations | Key Findings |
|---|---|---|---|---|
| SDS-PAGE | Separation by molecular weight in polyacrylamide gel | Porin expression profiling; relative quantification | Time-consuming; requires protein extraction | Detected OmpA, OmpK35, OmpK36 at ~35, ~36, ~37 kDa in K. pneumoniae |
| MALDI-TOF/MS | Mass spectrometry of extracted OMPs | Rapid porin detection; clinical laboratory application | Limited quantitative capability; extraction critical | Reliable detection of porins at expected m/z values; correlated with SDS-PAGE |
| Whole-Genome Sequencing | High-throughput DNA sequencing | Mutation detection; resistance gene identification | Cannot detect regulatory changes affecting expression | Identified truncations in ompK35 (13/26 isolates) and ompK36 (2/26 isolates) |
Functional Assessment of Porin Permeability
Understanding the functional consequences of porin deficiencies requires methodologies that directly measure membrane permeability and antibiotic transport:
Ion conductance measurements through reconstituted porins in lipid bilayers provide insights into channel properties. Studies on clinical OmpC variants showed that small ion unitary conductance did not significantly differ between mutants, indicating that resistance does not arise from major changes in pore size [67].
Fluorescent tracer uptake assays enable real-time assessment of porin permeability at the single-cell level. The fluorescent glucose analog 2NBDG, whose entry into E. coli is concentration- and time-dependent and mediated by porins, has been used to screen bacterial mutants for altered porin permeability [24]. Similarly, the fluorescent penicillin analog Bocillin FL and DNA stain Hoechst serve as tracers for antibiotic penetration [24].
Optogenetic approaches allow precise manipulation and monitoring of porin function. Expression of the light-activated proton pump ArchT in the inner membrane of E. coli enables selective acidification of the periplasm during fluorescence imaging, demonstrating that porin permeability decreases upon periplasmic acidification [24].
Diagram 2: Experimental workflow for comprehensive analysis of porin expression and function, integrating genomic, proteomic, and functional approaches.
Clinical Implications and Research Applications
Impact on Antibiotic Resistance
Porin deficiencies significantly contribute to resistance across multiple antibiotic classes, with particularly serious implications for last-resort treatments:
Carbapenem resistance frequently involves porin deficiencies working synergistically with β-lactamases. Porin loss or alteration reduces the carbapenem concentration in the periplasmic space, making the antibiotics more vulnerable to hydrolysis by β-lactamases such as extended-spectrum β-lactamases (ESBLs) and AmpC enzymes [64] [55]. This cooperation between reduced permeability and enzymatic degradation creates a formidable resistance phenotype.
Extended-drug resistance emerges when porin deficiencies combine with other resistance mechanisms. Clinical studies in Brazilian hospitals found that 74% of carbapenem-resistant P. aeruginosa isolates exhibited resistance to the novel combination drug imipenem-relebactam, largely attributable to porin loss and efflux pump overexpression rather than carbapenemase production [65]. Similarly, research on E. coli exposed to T-2 mycotoxin demonstrated that LamB downregulation contributed to extensively drug-resistant (XDR) phenotypes, particularly against last-resort antibiotics including cephalosporins, carbapenems, tigecycline, and colistin [70].
Fitness Costs and Virulence Alterations
The relationship between porin deficiencies and bacterial fitness presents a complex landscape with important clinical implications:
Metabolic trade-offs inevitably accompany porin deficiencies since these channels facilitate nutrient acquisition. However, bacteria implement compensatory mechanisms to mitigate these costs. In K. pneumoniae, porin loss triggers adaptations in other virulence factors; loss of a single porin reduces capsule production but increases LPS content, while dual porin loss significantly increases capsule production [68]. These adjustments help nullify fitness costs and maintain pathogenicity.
Virulence modulation varies depending on the specific porin alterations. K. pneumoniae isolates lacking OmpK35 alone or both OmpK35 and OmpK36 demonstrate reduced oxidative burst by macrophages and increased survival within phagocytes [68]. This enhanced immune evasion capacity illustrates how porin deficiencies can directly impact host-pathogen interactions beyond antibiotic resistance.
Contrary to the traditional view that resistance imposes fitness costs, some porin-deficient strains display maintained or even enhanced virulence. Studies of saturated mutant banks of P. aeruginosa and Acinetobacter baumannii suggest that porin-deficient strains may exhibit increased virulence in certain contexts [64] [55], presenting a worrying scenario for clinical management.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Porin Deficiency Research
| Reagent/Category | Specific Examples | Research Applications | Technical Function |
|---|---|---|---|
| Fluorescent Tracers | 2NBDG, Bocillin FL, Hoechst | Porin permeability assays; uptake kinetics | Monitor solute transport through porins in live cells |
| Genetically Encoded Sensors | pHluorin, pHuji, GINKO1, GINKO2, QuasAr2 | Real-time ion concentration; membrane potential | Measure periplasmic/cytoplasmic H+, K+; membrane voltage |
| Ionophores | CCCP, valinomycin | Manipulate ion gradients; membrane studies | Modulate H+ and K+ concentrations across membranes |
| Optogenetic Tools | ArchT proton pump | Controlled periplasmic acidification | Light-dependent manipulation of periplasmic pH |
| Porin Mutant Libraries | KEIO collection (E. coli) | High-throughput screening; functional genomics | Identify genes involved in porin regulation and function |
| Analytical Standards | ATCC strains (K. pneumoniae ATCC 13883, ATCC 700603) | Method validation; quality control | Reference materials for porin expression studies |
Porin deficiencies represent a sophisticated bacterial response to antibiotic pressure that significantly complicates the treatment of Gram-negative infections. Through downregulation, mutation, or complete loss of these outer membrane proteins, pathogens achieve reduced antibiotic permeability while implementing compensatory mechanisms to maintain fitness and virulence. The clinical relevance of these mechanisms is substantial, particularly in carbapenem-resistant Enterobacteriaceae and P. aeruginosa, which pose critical threats to global health.
Future research directions should focus on elucidating the precise structural changes that alter porin selectivity, developing methods to overcome permeability barriers, and exploiting the potential fitness costs of porin deficiencies for therapeutic benefit. The integrated experimental approaches outlined in this whitepaper—combining genomic, proteomic, and functional assessments—provide a roadmap for advancing our understanding of these complex resistance mechanisms. As drug development professionals confront the continuing challenge of antimicrobial resistance, a comprehensive understanding of porin biology will be essential for designing next-generation therapeutics capable of penetrating the Gram-negative outer membrane.
The outer membrane of Gram-negative bacteria serves as a formidable barrier against antimicrobial agents, primarily due to the presence of lipopolysaccharide (LPS) in its outer leaflet [71] [10]. LPS consists of three distinct regions: the O-antigen, core oligosaccharide, and lipid A [10] [72]. Lipid A, a glucosamine-based phospholipid embedded in the membrane, forms the fundamental scaffold of LPS and is essential for bacterial viability in nearly all Gram-negative species [10] [73]. This molecule is polyanionic, bearing multiple phosphate groups on its glucosamine disaccharide backbone, which creates a high negative charge density at the membrane surface [74] [10].
The negative charge of lipid A is crucial for the function of cationic antimicrobial peptides (AMPs) and last-resort antibiotics such as polymyxins [74] [75]. These positively charged antimicrobial molecules initially interact with the bacterial surface through electrostatic attractions to the phosphate groups of lipid A [74]. This interaction enables them to displace the divalent cations (Mg²⁺ and Ca²⁺) that stabilize the LPS layer, ultimately disrupting membrane integrity and causing cell death [10] [75]. This process is often termed "self-promoted uptake" [10] [75].
Given the critical role of lipid A in both membrane integrity and susceptibility to cationic antimicrobials, it is not surprising that bacteria have evolved sophisticated mechanisms to modify this molecule [74] [75]. These modifications represent a primary resistance mechanism against the innate immune response and last-resort antibiotics, framing lipid A as a key interface in the host-pathogen arms race and a significant factor in the context of outer membrane permeability and antibiotic resistance research [73] [72].
Mechanisms of Lipid A Modification
Bacteria employ two principal covalent modifications to alter the electrostatic properties of lipid A, thereby reducing its net negative charge and impairing cationic antibiotic binding. These modifications are mediated by distinct enzymatic systems that can be either chromosomally or plasmid-encoded.
4-Amino-4-deoxy-L-arabinose (L-Ara4N) Addition
The addition of the cationic sugar 4-amino-4-deoxy-L-arabinose (L-Ara4N) to one or both phosphate groups of lipid A is a widespread resistance mechanism among Gram-negative pathogens [74]. This modification is catalyzed by enzymes encoded by the arn (pmrE) operon, which is upregulated by two-component regulatory systems (TCSs) such as PmrA/PmrB and PhoP/PhoQ in response to environmental signals, including the presence of sublethal concentrations of cationic antimicrobials [76] [75]. The key enzyme ArnT (also known as PmrK) transfers the L-Ara4N moiety from the carrier molecule undecaprenyl-phosphate-α-L-Ara4N to the 1 or 4'-phosphate position of lipid A [74]. The arnA gene, which encodes the first functional enzyme in this modification pathway, has been experimentally validated as essential for the resistance phenotype [76].
Phosphoethanolamine (pEtN) Addition
The second major modification involves the transfer of a phosphoethanolamine (pEtN) group to the phosphate moieties of lipid A [74]. This reaction is primarily catalyzed by the enzyme EptA (also known as PmrC) in chromosome-encoded resistance [74] [75]. The eptA gene is similarly regulated by TCSs like PmrA/PmrB [75]. Notably, plasmid-encoded resistance is mediated by a family of mobilized colistin resistance (MCR) enzymes (MCR-1 to MCR-10) [74] [75]. These MCR enzymes are phosphoethanolamine transferases that catalyze the same biochemical reaction as EptA but are horizontally transmissible between bacterial strains and species, posing a significant public health threat [74].
Table 1: Key Lipid A Modification Enzymes and Their Features
| Enzyme | Gene(s) | Modification Added | Genetic Basis | Regulatory System |
|---|---|---|---|---|
| ArnT | arnT (part of arn operon) | L-Ara4N | Chromosomal | PmrA/PmrB, PhoP/PhoQ |
| EptA | eptA (pmrC) | pEtN | Chromosomal | PmrA/PmrB |
| MCR | mcr-1 to mcr-10 | pEtN | Plasmid | Constitutive (horizontal transfer) |
The net effect of both L-Ara4N and pEtN additions is a reduction of the net negative charge on the bacterial cell surface [74] [10]. This diminishes the initial electrostatic attraction between the membrane and cationic antimicrobials, leading to repulsion of molecules like polymyxin B and colistin and protecting bacteria from membrane disruption [74]. The following diagram illustrates the regulatory pathways and the resulting structural alterations to lipid A.
Diagram Title: Lipid A Modification Regulatory Pathways
Experimental Analysis and Detection
The study of lipid A modifications relies on a combination of genetic, phenotypic, and analytical techniques. The following section details key experimental protocols for investigating these resistance mechanisms.
Bacterial Survival Time-Kill Assay
This protocol determines the phenotypic tolerance of bacteria to cationic antibiotics or disinfectants after pre-exposure to sub-inhibitory concentrations.
Methodology:
- Culture Conditions: Grow bacterial isolates for 16 hours (to late stationary phase) under two conditions:
- Untreated control: Culture in standard growth medium (e.g., Luria Broth, LB).
- Pre-treated condition: Culture in medium containing a sub-inhibitory concentration (e.g., 30% v/v) of the stressor agent (e.g., disinfectant, low Mg²⁺) [76].
- Cell Preparation: Harvest 1 ml of culture by centrifugation. Resuspend the pellet in sterile phosphate-buffered saline (PBS) and adjust the suspension to a standardized optical density (e.g., OD600 of 0.1) [76].
- Challenge Exposure: Inoculate the washed cells into a lethal concentration of the antimicrobial agent (e.g., 100% v/v disinfectant, a high concentration of polymyxin) at a defined cell density (e.g., 1 × 10⁶ CFU/ml) [76].
- Viability Sampling: At designated time points, remove aliquots from the challenge solution and immediately transfer them into a neutralizing broth (e.g., Dey-Engley Neutralizing Broth) to stop antimicrobial action.
- Quantification: Perform serial dilution of the neutralized samples and plate on solid agar medium for colony-forming unit (CFU) counting after overnight incubation [76].
Interpretation: Enhanced survival of pre-treated bacteria compared to the untreated control indicates an inducible tolerance or resistance phenotype.
Genetic Analysis: Mutant Construction and Complementation
Defining the specific role of a gene in lipid A modification requires the creation and analysis of mutant strains.
Methodology:
- Strain Selection: Utilize a defined transposon mutant from a bacterial mutant library (e.g., P. aeruginosa PW7025 with a transposon insertion in the arnA gene) or construct a targeted gene deletion mutant via allelic exchange [76].
- Phenotypic Screening: Subject the mutant and the wild-type parent strain to the survival time-kill assay described above. A significant loss of the tolerance phenotype in the mutant (e.g., failure to survive in the disinfectant) implicates the target gene in the resistance mechanism [76].
- Complementation: Clone the wild-type gene into an expression plasmid and introduce it back into the mutant strain. Restoration of the wild-type phenotype confirms the gene's specific role [76].
Detection via Mass Spectrometry
Matrix-assisted laser desorption/ionization–time of flight–mass spectrometry (MALDI-TOF-MS) is a powerful tool for directly characterizing the structure of lipid A and its modifications.
Methodology:
- Lipid A Isolation: Extract total LPS from bacterial cells using a hot-phenol-water method. Cleave the ketosidic bond between lipid A and the core oligosaccharide by treating the purified LPS with mild acid hydrolysis to release lipid A [74].
- MALDI-TOF-MS Analysis:
- Sample Preparation: Mix the isolated lipid A with an appropriate matrix (e.g., 2,5-dihydroxybenzoic acid) and spot it on a target plate.
- Data Acquisition: Analyze the sample in negative ion mode, which is optimal for detecting the anionic lipid A molecules. The instrument measures the mass-to-charge ratio (m/z) of the native and modified lipid A ions [74].
- Data Interpretation: Identify the mass shifts corresponding to specific modifications. The addition of L-Ara4N results in a mass increase of +131 Da, while the addition of pEtN results in an increase of +123 Da [74]. The detection of these signature mass shifts provides direct evidence of lipid A modification.
Table 2: MALDI-TOF-MS Signature Ions for Lipid A Structures in Different Pathogens
| Bacterial Species | Native Lipid A m/z | Modification | Modified Lipid A m/z | Proposed Structure Change |
|---|---|---|---|---|
| Klebsiella pneumoniae | 1840 | + pEtN | 1963 (1840 + 123) | Plasmid-encoded (e.g., MCR) [74] |
| 1840 | + L-Ara4N | 1971 (1840 + 131) | Chromosome-encoded [74] | |
| Pseudomonas aeruginosa | 1462 | + L-Ara4N | 1593 (1462 + 131) | Single L-Ara4N addition [74] |
| 1462 | + 2 L-Ara4N | 1724 (1462 + 262) | Double L-Ara4N addition [74] | |
| Acinetobacter baumannii | 1910 | + pEtN | 2033 (1910 + 123) | Chromosome-encoded [74] |
Research Reagent Solutions
The following table compiles key reagents, materials, and tools essential for conducting research on lipid A modification pathways.
Table 3: Essential Research Reagents and Materials for Lipid A Studies
| Reagent / Material | Function / Application | Specific Example / Note |
|---|---|---|
| Dey-Engley Neutralizing Broth | Halts the killing action of antimicrobial agents (e.g., disinfectants, cationic antibiotics) during viability sampling in time-kill assays. | Critical for obtaining accurate colony counts post-exposure [76]. |
| Defined Bacterial Mutants | Links specific genes to phenotypic outcomes via gene-knockout studies. | Transposon mutant PW7025 (arnA::ISphoA/hah) used to validate arnA role in disinfectant tolerance [76]. |
| MALDI-TOF Mass Spectrometer | Detects and characterizes the precise structural modifications of isolated lipid A based on mass shifts. | Operated in negative ion mode for optimal analysis of lipid A phosphate groups [74]. |
| Sub-inhibitory Stressor Agents | Induces the expression of lipid A modification genes in vitro. | 30% (v/v) Opti-Free RepleniSH contact lens solution used to induce tolerance in P. aeruginosa [76]. Low Mg²⁺ or cationic peptides are also common inducers. |
| Anti-LPS Antibodies | Detects and quantifies LPS in biochemical transport assays (e.g., ELISA, Western Blot). | Used in vitro to monitor LPS release from transporter complexes in inhibition assays [71]. |
Implications for Drug Development and Concluding Perspectives
The study of lipid A modification pathways is more than an academic pursuit; it has direct implications for combating antibiotic resistance. Understanding these mechanisms informs the development of diagnostic tools and therapeutic strategies.
The ability to rapidly diagnose the mechanism of polymyxin resistance is crucial for infection control in clinical settings. MALDI-TOF-MS profiling of lipid A offers a rapid method to identify modifications, potentially distinguishing between chromosomal and the more concerning plasmid-encoded (MCR) resistance, which requires stricter containment measures [74]. Furthermore, the enzymes and regulatory systems involved in lipid A modification are themselves attractive drug targets. Inhibiting ArnT, EptA, or MCR enzymes, or the sensor kinases (e.g., PhoQ, PmrB) that control them, could re-sensitize resistant bacteria to cationic antibiotics and the host innate immune response [74] [75].
Recent advances highlight the potential of this approach. The inner membrane LPS transport complex, which must handle modified lipid A, has been successfully targeted by a new class of antibiotics, the macrocyclic peptides (e.g., Zosurabalpin) [71]. These drugs trap lipid A in its transporter, effectively stalling outer membrane biogenesis. Notably, the efficacy of these compounds can be affected by the structure of lipid A itself, such as the presence of an acyl chain added by LpxM, underscoring the interconnectedness of LPS biosynthesis, modification, and new antibiotic discovery [71].
In conclusion, lipid A modifications represent a primary and highly conserved resistance mechanism that directly impacts the efficacy of cationic antimicrobials by altering the physicochemical properties of the Gram-negative outer membrane. Continued research into the regulation, biochemistry, and functional consequences of these pathways is essential for developing novel countermeasures against multidrug-resistant Gram-negative infections.
The outer membrane (OM) of Gram-negative bacteria serves as a formidable barrier against antimicrobial agents, contributing significantly to intrinsic antibiotic resistance [10] [4]. This asymmetric bilayer, composed of phospholipids in the inner leaflet and lipopolysaccharides (LPS) in the outer leaflet, employs a sophisticated permeability system that restricts antibiotic penetration through porin-mediated diffusion for hydrophilic molecules and lipid-mediated pathways for hydrophobic compounds [10] [4]. When bacteria encounter environmental stresses—including antibiotics, host immune factors, or pH changes—they activate remodeling programs that alter OM structure and composition. While these adaptations enhance survival under immediate threat, they frequently incur substantial fitness costs that impact bacterial growth, virulence, and competitive ability [77] [78].
Understanding the trade-offs associated with OM remodeling provides crucial insights for developing novel antimicrobial strategies. This review examines the molecular mechanisms of OM adaptation, quantifies the associated physiological costs, and explores therapeutic approaches that exploit these vulnerabilities. The complex relationship between resistance and fitness represents a promising frontier in combating multidrug-resistant pathogens.
Molecular Mechanisms of Outer Membrane Remodeling
Lipopolysaccharide Modifications
Lipopolysaccharide serves as a primary component of the outer leaflet of the OM, and its modification represents a fundamental resistance mechanism. Under conditions of cationic antimicrobial peptide exposure or magnesium limitation, Salmonella enterica and related pathogens activate the PhoP/Q and PmrA/B two-component systems [77]. These regulators induce covalent modifications to the lipid A moiety of LPS, including the addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) and phosphoethanolamine (pEtN) groups to phosphate moieties, and palmitoylation of acyl chains [77]. These alterations reduce the net negative charge of the membrane, decreasing affinity for cationic antimicrobial peptides and promoting membrane stability in low-divalent cation environments [77].
Table 1: LPS Modifications and Their Functional Consequences
| Modification Type | Inducing Conditions | Enzymes Involved | Impact on Membrane Properties |
|---|---|---|---|
| l-Ara4N addition to lipid A phosphates | Low Mg²⁺, acidic pH, cationic antimicrobial peptides | PmrC, PbgP | Reduces net negative charge, decreases cationic antimicrobial peptide binding |
| pEtN addition to lipid A phosphates | Low Mg²⁺, acidic pH | CptA | Neutralizes phosphate negative charges, stabilizes LPS layer |
| Palmitoylation at position 2 of lipid A | Cationic antimicrobial peptides, low Mg²⁺ | PagP | Increases hydrophobic interactions, enhances bilayer stability |
| Core oligosaccharide truncation | Mutations in LPS biosynthesis genes | Wa* proteins | Increases membrane permeability to hydrophobic agents |
Outer Membrane Protein Alterations
The protein composition of the OM undergoes significant changes during adaptive remodeling. Bacteria can downregulate general diffusion porins such as OmpF and OmpC, reducing the penetration of hydrophilic antibiotics like β-lactams [10] [4]. Conversely, stress conditions may upregulate specific transporters for nutrient acquisition or efflux pump components that export toxic compounds. The differential expression of these outer membrane proteins (OMPs) represents a strategic reallocation of resources that balances protection with physiological requirements [10] [4].
Vesiculation as a Remodeling Mechanism
Gram-negative bacteria constitutively shed outer membrane vesicles (OMVs), and this process accelerates during envelope stress [77]. OMVs facilitate the selective removal of misfolded proteins and potentially detrimental LPS species from the OM. Research demonstrates that palmitoylated lipid A species incorporate into OMVs at disproportionately high rates, suggesting vesiculation serves as a quality control mechanism to maintain optimal membrane fluidity and integrity under changing conditions [77].
Diagram Title: OM Remodeling Triggered by Environmental Stress
Quantitative Analysis of Fitness Costs
The physiological burdens associated with OM remodeling manifest across multiple cellular processes. The following table synthesizes experimental measurements of these fitness costs from representative studies.
Table 2: Experimentally Determined Fitness Costs of OM Remodeling
| Modification Type | Experimental Model | Growth Rate Reduction | Virulence Attenuation | Competitive Index Deficit | Reference |
|---|---|---|---|---|---|
| l-Ara4N/pEtN lipid A modifications | Salmonella enterica serovar Typhimurium | 15-25% in minimal media | 10-100x LD₅₀ increase in murine model | 2-5x after 20 generations | [77] |
| Palmitoylated lipid A incorporation into OMVs | Salmonella enterica serovar Typhimurium | Not significant | Not directly measured | 30% enrichment in OMV fraction | [77] |
| Porin downregulation (OmpF/OmpC) | Escherichia coli | 5-15% in nutrient-rich media | Variable depending on infection site | 1.5-3x in mixed culture | [10] [4] |
| Efflux pump overexpression | Pseudomonas aeruginosa | 10-20% in absence of antibiotics | Context-dependent | 2-4x in antibiotic-free medium | [79] [80] |
The metabolic costs of resistance mechanisms extend beyond growth parameters. Bacteria with modified LPS exhibit increased sensitivity to detergents and certain antibiotics, reflecting the delicate balance between resistance to specific threats and overall membrane integrity [77]. Mutations that confer phage resistance frequently involve alterations in outer membrane proteins that also function in nutrient import, creating antagonistic pleiotropy where resistance comes at the expense of metabolic efficiency [78].
Experimental Approaches for Assessing OM Remodeling
LPS Analysis Protocol
The comprehensive characterization of LPS modifications employs a multidisciplinary approach:
Bacterial Culture under Inducing Conditions: Grow Salmonella enterica serovar Typhimurium to mid-logarithmic phase (OD₆₀₀ ≈ 0.6) in neutral, high-Mg²⁺ medium (pH 7.6, 10 mM MgSO₄). Harvest cells by gentle centrifugation and resuspend in fresh medium under either the same conditions or shift to mildly acidic, low-Mg²⁺ medium (pH 5.8, 10 µM MgSO₄) to activate PhoP/Q and PmrA/B systems [77].
Lipid A Extraction: Hydrolyze purified LPS samples in mild acid conditions (10 mM sodium acetate, pH 4.5, 1% SDS) at 100°C for 1 hour. Extract released lipid A using chloroform-methanol-water mixture [77].
Mass Spectrometry Analysis: Analyze lipid A specimens using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry in negative ion mode. Compare mass profiles to identify covalent modifications (l-Ara4N, pEtN, palmitoylation) based on mass shifts [77].
OMV Isolation and Analysis: Separate OMVs from culture supernatants by sequential centrifugation and ultracentrifugation. Analyze OMV lipid A content using the same mass spectrometry approaches to determine modification enrichment factors compared to whole-cell extracts [77].
Fitness Cost Assessment Methods
Quantifying the physiological trade-offs of OM remodeling requires complementary assays:
Growth Kinetics Monitoring: Measure bacterial growth rates in controlled environments using optical density (OD₆₀₀) measurements or direct cell counting. Compare maximal growth rates and carrying capacities between modified and wild-type strains [77].
Competition Assays: Co-culture modified and wild-type strains in a 1:1 ratio for approximately 20 generations. Determine competitive indices by plating on selective media or using differential staining and fluorescence-activated cell sorting [77] [78].
Virulence Assessment: Evaluate pathogenicity using appropriate animal models (typically murine infection models for Salmonella). Compare median lethal dose (LD₅₀) values, bacterial burden in target organs, and histopathological changes [77].
Metabolic Profiling: Utilize transcriptomics, proteomics, and metabolomics to identify pathway alterations associated with resistance mechanisms. Measure nutrient uptake rates using radiolabeled compounds or fluorescent analogs [78].
Diagram Title: Experimental Workflow for OM Remodeling Analysis
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for OM Remodeling Research
| Reagent/Category | Specific Examples | Research Application | Technical Notes | |
|---|---|---|---|---|
| Defined Growth Media | N-minimal medium (pH 5.8 vs 7.6, 10 µM vs 10 mM Mg²⁺) | Induction of PhoP/Q and PmrA/B systems | Carefully control divalent cation concentrations using chelators | [77] |
| Mass Spectrometry Standards | Synthetic lipid A standards, l-Ara4N-modified lipid A | Quantification of LPS modifications | Use internal standards for accurate relative quantification | [77] |
| OMV Isolation Reagents | Ultracentrifugation equipment, density gradient media | Separation of OMVs from culture supernatant | Include protease inhibitors to prevent degradation | [77] |
| Antibiotic Susceptibility Testing | Cationic antimicrobial peptides (e.g., polymyxin B), β-lactams | Functional assessment of permeability barriers | Use standardized inoculum for reproducibility | [10] [77] |
| Genetic Tools | Knockout mutants (e.g., ∆pmrA, ∆phoP), complementation vectors | Establish causal relationships between genes and phenotypes | Use in-trans complementation to confirm specificity | [77] |
| OM Permeability Probes | Nitrocefin, N-phenyl-1-naphthylamine (NPN), ethidium bromide | Quantitative assessment of OM permeability | NPN assay measures uptake of hydrophobic compounds | [10] [4] |
Therapeutic Implications and Future Directions
The fitness trade-offs associated with OM remodeling present attractive opportunities for therapeutic intervention. The concept of "collateral sensitivity"—where resistance to one antimicrobial increases susceptibility to another—informs strategic combination therapies [78]. For instance, bacteria with modified LPS that resist cationic antimicrobial peptides may exhibit enhanced sensitivity to hydrophobic antibiotics due to altered membrane fluidity properties [77] [78].
Bacteriophage therapy strategically exploits these trade-offs. Phages that target specific outer membrane structures select for resistant mutants with receptor modifications that frequently impair nutrient uptake or restore antibiotic sensitivity [78]. This evolutionary steering approach uses phages as biological tools to manipulate bacterial populations toward more susceptible states.
Advanced understanding of OM remodeling mechanisms informs drug development targeting resistance itself. Inhibitors of LPS modification enzymes (e.g., L-Ara4N transferases) could prevent resistance development without directly killing bacteria, potentially reducing selective pressure for future resistance [77] [80]. Similarly, small molecules that disrupt the regulation of efflux pumps or porin expression may restore activity of existing antibiotics [79] [80].
The integration of artificial intelligence and machine learning in antibiotic discovery accelerates the identification of compounds that exploit fitness trade-offs [79] [80]. These approaches analyze complex datasets to predict resistance evolution and design antimicrobials with a higher barrier to resistance development.
Outer membrane remodeling represents a sophisticated bacterial adaptation to environmental threats, but one that inevitably extracts physiological costs. The precise molecular characterization of these trade-offs, combined with innovative therapeutic strategies that exploit them, offers promising avenues to address the escalating crisis of antibiotic resistance. Future research should focus on quantifying these fitness costs across diverse bacterial species and infection environments, developing high-throughput screening methods for collateral sensitivity, and advancing combination therapies that leverage evolutionary principles. Such approaches will enhance our ability to outmaneuver bacterial adaptation and preserve the efficacy of antimicrobial agents.
The escalating global threat of antimicrobial resistance necessitates innovative strategies to rejuvenate the efficacy of existing antibiotics. The synergy between the bacterial outer membrane permeability barrier and active efflux pumps constitutes a formidable defense mechanism, rendering many potent antibiotics ineffective. This whitepaper delineates the critical role of efflux pump inhibition (EPI) as a strategy to overcome multidrug resistance, particularly in the context of compromised outer membrane integrity. We present quantitative evidence of synergy between permeabilizing agents and EPIs, detailed experimental methodologies for screening such synergistic interactions, and an overview of emerging inhibitor classes. The data underscore that combining permeability-increasing compounds with EPIs can dramatically lower the required inhibitory concentration of both components, effectively resensitizing resistant bacteria to conventional antibiotics and presenting a viable path for therapeutic intervention.
The intrinsic resistance of Gram-negative bacteria to a broad spectrum of antibiotics is largely attributed to the synergistic action of two key cellular features: a formidable outer membrane and a network of active efflux pumps [10] [81]. The outer membrane, with its asymmetric bilayer of phospholipids and lipopolysaccharides (LPS), serves as a potent permeability barrier, significantly limiting the intracellular accumulation of antibiotics [10]. Lipopolysaccharides in the outer leaflet, with their strong lateral interactions and low fluidity, create a highly impermeable hurdle, particularly for hydrophobic compounds [10] [81].
Simultaneously, bacteria employ energy-dependent efflux pumps that actively recognize and extrude a wide range of structurally diverse antibiotics from the cell, further reducing the intracellular drug concentration to sub-lethal levels [82]. These membrane transport proteins are categorized into several families based on their structure and energy source, including the Resistance Nodulation Division (RND) superfamily, which are particularly potent in mediating antibiotic resistance in Gram-negative bacteria [82]. The interplay between these two systems creates a highly effective defense: a drug that successfully traverses the outer membrane may still be expelled before reaching its intracellular target [37] [10]. Overcoming this dual barrier is paramount to combating multidrug-resistant infections.
The Scientific Basis: Mechanisms and Synergistic Potential
Efflux Pump Families and Their Roles
Efflux pumps are broadly classified into primary active transporters, which utilize ATP hydrolysis, and secondary active transporters, which leverage the proton motive force (PMF). The major families include:
- ATP-binding Cassette (ABC) Superfamily: These primary transporters utilize ATP hydrolysis to translocate substrates. They consist of two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs) [82].
- Resistance Nodulation Division (RND) Superfamily: Often forming tripartite complexes (e.g., AcrAB-TolC in E. coli, MexAB-OprM in P. aeruginosa), these pumps are major contributors to multidrug resistance in Gram-negative bacteria. They function as proton-substrate antiporters [82].
- Major Facilitator Superfamily (MFS): This is the largest superfamily of secondary active transporters, which utilize the proton motive force to drive substrate efflux [82].
The Permeability-Inhibition Synergy
The conceptual breakthrough lies in simultaneously targeting the physical barrier (outer membrane) and the active removal system (efflux pumps). Permeabilizing agents, such as polymyxin B nonapeptide (PMBN), disrupt the dense packing of LPS in the outer membrane, creating pathways for increased antibiotic influx [10] [83]. When this is combined with an EPI that blocks the efflux of the antibiotic, the net intracellular accumulation of the antibiotic increases dramatically, restoring its efficacy [83].
Research on P. aeruginosa strains overexpressing the MexAB-OprM efflux system demonstrates that PMBN synergizes with EPIs like PAβN and NMP. This synergy drastically reduces the amount of EPI required for antibiotic sensitization, potentially mitigating toxicity concerns associated with high doses of EPIs [83]. The underlying mechanism for some EPIs involves altering the proton motive force (PMF), diminishing membrane potential (ΔΨ) and inhibiting ATP production, thereby crippling the energy source for many efflux pumps [84].
Diagram 1: The dual barrier mechanism and intervention strategy. The outer membrane limits antibiotic influx, while efflux pumps actively expel drugs that enter. Combining a membrane permeabilizer and an efflux pump inhibitor synergistically increases intracellular antibiotic concentration.
The synergistic effect of combining permeabilizing agents with EPIs is demonstrated by significant reductions in the Minimum Inhibitory Concentration (MIC) of antibiotics against multidrug-resistant strains. The data is quantified using the Fractional Inhibitory Concentration Index (FICI), where FICI ≤ 0.5 indicates synergy.
Table 1: Synergistic Effect of PMBN and EPIs on Antibiotic Efficacy in P. aeruginosa LC1-6
| Antibiotic | MIC Alone (µg/mL) | EPI Used | Combination MIC (µg/mL) | Fold Reduction in MIC | FICI Value |
|---|---|---|---|---|---|
| Azithromycin | 128 | PAβN | 0.06 | 2,133 | 0.002 |
| Ceftazidime | 128 | PAβN | 0.5 | 256 | 0.007 |
| Levofloxacin | 8 | PAβN | 0.06 | 133 | 0.008 |
| Piperacillin | 256 | PAβN | 2 | 128 | 0.008 |
| Aztreonam | 32 | PAβN | 2 | 16 | 0.07 |
| Azithromycin | 128 | NMP | 1 | 128 | 0.01 |
Source: Adapted from [83]. Combinations included a fixed, sub-inhibitory concentration of PMBN.
Table 2: Activity of Repurposed Antifungals as Efflux Pump Inhibitors in S. aureus
| Efflux Pump Inhibitor | Key Mechanism | Impact on Antibiotic MIC | In Vivo Model Effect |
|---|---|---|---|
| Sertaconazole | Alters PMF; reduces membrane potential (ΔΨ) without changing efflux rate. | Enhanced efficacy of norfloxacin, cefotaxime, and moxifloxacin. | Significantly lowered bacterial load in a murine skin infection model. |
| Oxiconazole | Alters PMF; reduces both efflux pump activity and efflux rate. | Enhanced efficacy of norfloxacin, cefotaxime, and moxifloxacin. | Significantly lowered bacterial load in a murine skin infection model. |
Source: Summarized from [84]. Both drugs showed minimal cytotoxicity toward mammalian cells.
Experimental Protocols: Methodologies for Key Assays
Checkerboard Synergy Assay for EPI and Permeabilizer Screening
This protocol is designed to identify and quantify synergy between an antibiotic, an EPI, and a membrane permeabilizer like PMBN [83].
Bacterial Strain and Inoculum Preparation:
- Use a standardized inoculum from a mid-logarithmic phase culture of the target bacterium (e.g., P. aeruginosa LC1-6). Adjust the turbidity to approximately 1-5 x 10^8 CFU/mL and further dilute in the appropriate broth medium to a working concentration of ~5 x 10^5 CFU/mL.
Checkerboard Microdilution Setup:
- Prepare a two-dimensional checkerboard in a 96-well microtiter plate. Serially dilute the antibiotic along the rows (e.g., two-fold dilutions). Serially dilute the EPI (e.g., PAβN or NMP) along the columns.
- To all wells, add a fixed, sub-inhibitory concentration of the membrane permeabilizer (e.g., 1-2 µg/mL of PMBN). This creates a matrix where each well contains a unique combination of antibiotic, EPI, and a constant level of permeabilizer.
- Include control wells for growth (no drugs), antibiotic alone, EPI alone, and PMBN alone.
Incubation and MIC Determination:
- Inoculate each well with the prepared bacterial suspension.
- Incubate the plate under optimal conditions for the bacterial strain (e.g., 35±2°C for 18-20 hours).
- Determine the Minimum Inhibitory Concentration (MIC) for the antibiotic in the presence of the fixed PMBN and varying EPI concentrations. The MIC is defined as the lowest concentration that prevents visible growth.
Data Analysis and FICI Calculation:
- Calculate the Fractional Inhibitory Concentration Index (FICI) to quantify synergy. The FICI for a triple combination (Antibiotic + EPI + Permeabilizer) can be interpreted as:
FICI = (MIC_antibiotic in combination / MIC_antibiotic alone) + (MIC_EPI in combination / MIC_EPI alone) - Interpretation: FICI ≤ 0.5 indicates synergy; >0.5 to ≤4.0 indicates no interaction (additivity/indifference); and >4.0 indicates antagonism.
- Calculate the Fractional Inhibitory Concentration Index (FICI) to quantify synergy. The FICI for a triple combination (Antibiotic + EPI + Permeabilizer) can be interpreted as:
Direct Quantification of Antibiotic Accumulation via LC-MS
To directly confirm that EPIs increase intracellular antibiotic concentration, accumulation assays can be performed [37].
Sample Preparation:
- Grow bacterial cultures (e.g., Mycobacterium abscessus or target Gram-negative strains) to mid-log phase.
- Expose the bacterial cells to the antibiotic of interest in the presence and absence of a pre-optimized concentration of an EPI.
- Incubate for a defined period (e.g., 4 hours) to allow for accumulation.
Cell Harvesting and Washing:
- At the end of the incubation, rapidly centrifuge the cells to pellet them.
- Wash the pellet with an appropriate buffer (e.g., phosphate-buffered saline, pH 7.0) to remove any extracellular antibiotic adhered to the cell surface. This step is critical for accurate measurement.
Cell Lysis and Metabolite Extraction:
- Lyse the bacterial cells using a robust method such as bead-beating or sonication in a suitable extraction solvent (e.g., a mixture of methanol and water).
- Remove cell debris by centrifugation and collect the supernatant containing the intracellular metabolites and the accumulated antibiotic.
LC-MS Analysis and Quantification:
- Analyze the extracted supernatants using Liquid Chromatography-Mass Spectrometry (LC-MS).
- Use a panel of antibiotic standards to generate a calibration curve for absolute or relative quantification.
- Compare the peak areas or concentrations of the antibiotic in samples treated with EPIs versus untreated controls. A statistically significant increase in the EPI-treated samples confirms efflux pump inhibition and enhanced drug accumulation [37].
Diagram 2: Experimental workflow for quantifying intracellular antibiotic accumulation via LC-MS. This protocol directly measures the effect of EPIs on drug retention inside bacterial cells.
Emerging Inhibitors and Research Tools
Table 3: Key Reagents and Tools for Efflux Pump and Permeability Research
| Tool / Reagent | Function / Description | Example Use |
|---|---|---|
| PAβN (Phe-Arg-β-naphthylamide) | Broad-spectrum EPI for Gram-negative bacteria; substrate of RND pumps like MexB and AcrB. | Synergy studies with permeabilizers and antibiotics against P. aeruginosa and E. coli [83]. |
| NMP (1-(1-Naphthylmethyl)-piperazine) | Structurally distinct EPI from PAβN; also acts as an efflux pump substrate. | Used to confirm synergy is not limited to a single EPI class [83]. |
| PMBN (Polymyxin B Nonapeptide) | Membrane permeabilizer derived from polymyxin B; disrupts outer membrane with lower toxicity. | Lowers the effective concentration of EPIs required for antibiotic sensitization [83]. |
| Repurposed Azoles (Sertaconazole, Oxiconazole) | FDA-approved antifungal drugs identified as potent EPIs in S. aureus. | Restore susceptibility to fluoroquinolones and other antibiotics; promising for drug repurposing [84]. |
| Plant-Derived Compounds (Berberine, Palmatine, Curcumin) | Natural products with documented EPI and Sortase A inhibitory activity. | Potential as antibiotic potentiators and components of combination therapy; often have dual mechanisms [85]. |
| Bac-EPIC Web Server | An in silico platform for predicting EPI compounds targeting the AcrAB-TolC pump in E. coli. | Aids in early-stage drug discovery by screening chemical structures for potential EPI activity [86]. |
The strategic inhibition of efflux pumps, particularly when combined with agents that compromise the integrity of the outer membrane, represents a powerful and clinically viable approach to countering multidrug-resistant bacterial infections. Quantitative data unequivocally demonstrates that this synergy can reduce the MIC of legacy antibiotics by several orders of magnitude, effectively resensitizing even highly resistant strains. The ongoing identification of novel EPIs—from synthetic compounds to repurposed drugs and natural products—provides a rich pipeline for therapeutic development. Future research must focus on optimizing these combinations for clinical use, thoroughly assessing their safety and pharmacokinetics, and deploying advanced tools for inhibitor discovery. Overcoming the dual barrier of membrane permeability and active efflux is not just a promising research avenue but a critical imperative for preserving the efficacy of our antimicrobial arsenal.
The rapid rise of antimicrobial resistance (AMR) represents one of the most pressing global public health threats of the 21st century. According to the World Health Organization (WHO), one in six laboratory-confirmed bacterial infections in 2023 were resistant to antibiotic treatments, with resistance rising in over 40% of monitored pathogen-antibiotic combinations [87]. Gram-negative bacteria—particularly Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa—pose a formidable challenge due to their complex cell envelope structure, which includes an asymmetric outer membrane that serves as a formidable permeability barrier [3] [5].
The outer membrane of Gram-negative bacteria, with its lipopolysaccharide (LPS)-rich outer leaflet, provides intrinsic resistance to many hydrophobic antibiotics and toxins [5]. Hydrophilic antibiotics primarily cross this barrier through porin channels, but bacteria can develop resistance by modifying these channels or their regulation [3]. This architectural defense has rendered many conventional antibiotics ineffective, necessitating innovative approaches to restore their efficacy.
Combinatorial therapies that pair membrane-active agents with conventional antibiotics represent a promising strategy to overcome this bacterial defense system. By disrupting membrane integrity, membrane-active agents can increase antibiotic penetration and potentially circumvent established resistance mechanisms [88] [89]. This comprehensive review examines the scientific basis, experimental evidence, and practical methodologies for developing such combination therapies, providing researchers with both theoretical foundations and actionable protocols.
Membrane Permeability: The Gram-Negative Barrier
Molecular Organization of the Outer Membrane
The Gram-negative outer membrane exhibits unique structural characteristics that contribute to its function as a protective barrier:
- Asymmetric Bilayer Structure: The outer membrane consists of an inner leaflet composed of phospholipids and an outer leaflet dominated by lipopolysaccharide (LPS) molecules [5].
- LPS Architecture: Each LPS molecule contains three domains: lipid A (a glucosamine-based phospholipid embedded in the membrane), a core oligosaccharide, and the O-antigen polysaccharide that extends extracellularly [5].
- Porin Channels: General diffusion porins such as OmpF, PhoE, and OmpC form trimeric β-barrel structures that create water-filled channels for hydrophilic molecule passage [5].
Permeability Pathways and Resistance Mechanisms
Antibiotics primarily traverse the outer membrane through two principal pathways:
- Lipid-Mediated Pathway: Hydrophobic antibiotics diffuse through the membrane bilayer itself, with efficacy dependent on LPS composition and fluidity [3].
- Porin-Mediated Pathway: Hydrophilic antibiotics utilize water-filled porin channels, with size exclusion limits (typically <600 Da) and charge selectivity influencing permeability [3] [5].
Bacteria employ multiple resistance strategies targeting these pathways, including LPS modifications to reduce hydrophobic compound uptake, porin downregulation to limit hydrophilic antibiotic entry, and efflux pump overexpression to actively remove compounds that successfully penetrate the membrane [3] [5].
Diagram 1: Gram-negative cell envelope structure and antibiotic penetration pathways. Membrane-active agents target both LPS and porin components to enhance permeability.
High-Throughput Screening Platforms for Combination Discovery
DropArray Technology for Large-Scale Combination Screening
Recent advances in high-throughput screening have enabled the systematic evaluation of millions of drug combinations. The DropArray platform represents a significant technological innovation, using nanoliter-scale miniaturization and random self-assembly of droplet combinations on microwell array chips to overcome the limitations of conventional plate-based methods [88].
Key Methodology:
- Platform Design: DropArray utilizes a droplet-in-microarray technology that enables testing of over 1.3 million unique strain-antibiotic-compound combinations [88].
- Barcoding System: A fluorescent dye-based barcoding strategy identifies antibiotic and compound inputs in each assay [88].
- Bacterial Load Measurement: Engineered bacterial strains expressing GFP allow fluorescence-based quantification of bacterial biomass reduction [88].
In a landmark study applying this technology, researchers screened over 30,000 compounds in combination with up to 22 antibiotics against six Gram-negative ESKAPE pathogen strains. This approach identified P2-56, a small molecule that synergized with rifampin against A. baumannii and K. pneumoniae [88]. A derivative compound, P2-56-3, demonstrated enhanced potency and was found to compromise outer membrane integrity, facilitating rifampin penetration [88].
Artificial Intelligence-Driven Combination Optimization
The IDentif.AI-AMR platform represents an alternative approach that uses artificial intelligence to efficiently navigate combination space without exhaustive experimental testing [90].
Platform Workflow:
- Experimental Design: Tests a carefully selected subset of combinations using Orthogonal Array Composite Design (OACD) to maximize information gain [90].
- Mathematical Modeling: Employs a second-order quadratic model to correlate drug combinations with efficacy outcomes [90].
- Optimization: Rapidly pinpoints optimal combinations from the parameter space without testing all possible combinations [90].
In practice, this platform identified effective combinations against extremely drug-resistant A. baumannii from a pool of nine FDA-approved drugs. Notably, the cefiderocol/polymyxin B/rifampicin combination achieved 92.23 ± 11.89% bacterial inhibition, while the polymyxin B/rifampicin pair demonstrated broad-spectrum efficacy across multiple clinical isolates [90].
Diagram 2: Complementary approaches for discovering synergistic antibiotic combinations. High-throughput screening (top) and AI-driven optimization (bottom) represent two powerful methodologies.
Experimental Evidence and Efficacy Data
Documented Synergistic Combinations
Table 1: Efficacy of documented synergistic combinations against Gram-negative pathogens
| Combination | Pathogen | Efficacy/Interaction | Proposed Mechanism | Reference |
|---|---|---|---|---|
| P2-56-3 + Rifampin | A. baumannii, K. pneumoniae | Synergistic | Compromises outer membrane integrity; genetic dependency on lpt genes | [88] |
| Polymyxin B + Rifampicin | A. baumannii (clinical isolates) | 92.23% inhibition; strong synergy | Membrane disruption enhancing intracellular antibiotic access | [90] |
| Cefiderocol + Polymyxin B + Rifampicin | A. baumannii | 92.23 ± 11.89% inhibition | Combined membrane permeabilization and iron transport-mediated uptake | [90] |
| Ampicillin-sulbactam + Cefiderocol | A. baumannii | 93.89 ± 5.95% inhibition | β-lactamase inhibition with enhanced penetration | [90] |
| Antimicrobial Peptides + Conventional Antibiotics | Multiple WHO priority pathogens | Variable synergy | Membrane disruption, efflux pump inhibition, biofilm penetration | [89] |
Mechanisms of Synergy at the Molecular Level
The synergistic activity between membrane-active agents and conventional antibiotics operates through several distinct mechanisms:
- Outer Membrane Disruption: Compounds like P2-56-3 compromise the integrity of the outer membrane, as demonstrated by hypersensitivity in mutants with compromised lipooligosaccharide transport systems (lptA-D, lptFG) [88].
- Efflux Pump Interference: Certain combinations counteract resistance-nodulation-division (RND) efflux pump activity, which normally exports antibiotics from the cell [88].
- Biofilm Penetration: Antimicrobial peptides (AMPs) can enhance antibiotic efficacy against biofilm-embedded bacteria by improving penetration through the extracellular matrix [89].
- Intracellular Delivery: Some membrane-active agents facilitate the delivery of antibiotics to intracellular bacterial compartments [89].
Contradictory Evidence and Limitations
Despite promising results, the literature reveals contradictory findings that highlight the complexity of combination approaches:
- Lack of Universal Synergy: A systematic study measuring Fractional Inhibitory Concentration (FIC) for combinations of membrane-permeabilizing antimicrobial peptides with conventional antibiotics found no statistically significant synergy in any of the 40 pairwise combinations tested [91].
- Mechanistic Limitations: Large-scale membrane disruption and permeabilization alone may not be sufficient to drive synergistic interactions with conventional antibiotics [91].
- Context-Dependent Activity: The same membrane-active agent may show different synergistic potential depending on the specific antibiotic partner, bacterial strain, and environmental conditions [91].
Experimental Protocols and Methodologies
High-Throughput DropArray Screening Protocol
Objective: Systematically identify synergistic compound-antibiotic combinations against Gram-negative pathogens.
Materials:
- DropArray microarray chips
- Bacterial strains engineered for GFP expression
- Compound libraries (30,000+ compounds)
- Antibiotic panels (up to 22 antibiotics at 2-3 concentrations)
- Fluorescent barcoding dyes
- Automated imaging system
Procedure:
- Prepare bacterial suspension in appropriate growth medium at standardized density.
- Dispense nanoliter-scale droplets of bacteria, compounds, and antibiotics onto the DropArray chip.
- Allow random self-assembly of droplet combinations in microwells.
- Use fluorescent barcoding to identify the contents of each combination.
- Incubate under appropriate conditions (e.g., 37°C for 20 hours).
- Measure bacterial growth via GFP fluorescence intensity.
- Calculate Z-prime scores using bacteria-only and media-only controls as positive and negative controls, respectively.
- Analyze combination effects using appropriate synergy models.
- Validate hits through secondary assays [88].
Fractional Inhibitory Concentration (FIC) Determination
Objective: Quantitatively measure interactions between membrane-active agents and antibiotics.
Materials:
- 96-well microtiter plates
- Logarithmic-phase bacterial cultures
- Serial dilutions of test compounds
- Spectrophotometer for optical density measurements
Procedure:
- Prepare serial dilutions of membrane-active agent and antibiotic in a checkerboard pattern.
- Inoculate each well with standardized bacterial suspension (e.g., 4×10^5 CFU/mL).
- Incubate plates under appropriate conditions (e.g., 37°C for 16-20 hours).
- Measure bacterial growth via optical density at 600 nm.
- Determine Minimum Inhibitory Concentration (MIC) for each agent alone and in combination.
- Calculate FIC indices using the formula:
- Interpret results:
- FIC ≤ 0.5: Synergy
- 0.5 < FIC ≤ 4: Additivity or indifference
- FIC > 4: Antagonism [91]
Mechanism Elucidation Through Genetic Screens
Objective: Identify genetic dependencies of synergistic combinations.
Materials:
- CRISPRi bacterial libraries
- Conditional expression systems
- Phenotypic microarrays
- Western blot equipment
Procedure:
- Generate CRISPRi knockdown strains targeting potential pathway genes.
- Expose knockdown strains to synergistic combinations at sub-inhibitory concentrations.
- Compare growth inhibition between knockdown and wild-type strains.
- Identify hypersensitive mutants indicating genetic dependencies.
- Validate findings through complementation assays.
- Perform phenotypic assays to confirm mechanism (e.g., membrane permeability assays, efflux pump activity measurements) [88].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key research reagents for investigating antibiotic-membrane agent combinations
| Reagent/Category | Specific Examples | Function/Application | Reference |
|---|---|---|---|
| High-Throughput Screening Platforms | DropArray, Orthogonal Array Composite Design (OACD) | Enables large-scale combination screening with minimal reagent use | [88] [90] |
| Membrane-Active Compounds | P2-56-3, Polymyxin B, Antimicrobial Peptides (AMPs) | Disrupt outer membrane integrity to enhance antibiotic penetration | [88] [89] [90] |
| Synergy Quantification Tools | Fractional Inhibitory Concentration (FIC), Z-prime scores | Provides standardized metrics for evaluating combination effects | [88] [91] |
| Genetic Tools | CRISPRi knockdown strains, Transposon mutant libraries | Elucidates mechanisms and genetic dependencies of synergistic combinations | [88] |
| Permeability Assays | SYTOX Green uptake, NPN assay, ONPG hydrolysis | Measures outer membrane disruption and compound penetration | [91] |
| Clinical Isolate Panels | ESKAPE pathogen collections, Carbapenem-resistant strains | Validates combination efficacy against clinically relevant strains | [88] [90] |
Combinatorial approaches pairing membrane-active agents with conventional antibiotics represent a promising strategy to overcome the permeability barriers of Gram-negative pathogens. High-throughput screening platforms and AI-driven optimization methods have accelerated the discovery of effective combinations, such as P2-56-3 with rifampin and polymyxin B with rifampicin, which demonstrate significant efficacy against multidrug-resistant strains.
Future research directions should focus on several key areas:
- Mechanism Elucidation: Deeper investigation into the molecular pathways underlying synergistic interactions.
- Resistance Mitigation: Understanding how combination therapies affect the development and selection of resistant subpopulations.
- Clinical Translation: Addressing formulation challenges and pharmacokinetic optimization for combination regimens.
- Expanded Screening: Applying high-throughput platforms to broader compound libraries and additional pathogen species.
As antibiotic resistance continues to escalate, combinatorial approaches that target both membrane permeability and intracellular processes offer a promising path forward for maintaining efficacy against Gram-negative pathogens. The integration of advanced screening technologies with mechanistic studies provides a robust framework for developing the next generation of antimicrobial therapies.
Pathogen-Specific Barriers: Comparative Permeability and Resistance Across Species
The outer membrane (OM) of Gram-negative bacteria serves as a primary barrier against antibiotics, with its permeability characteristics varying significantly across species. Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii represent a spectrum of OM impermeability, with P. aeruginosa and A. baumannii exhibiting notably lower permeability that contributes to their intrinsic multidrug resistance. This technical review examines the structural basis for these differences, highlighting species-specific porin profiles, lipopolysaccharide (LPS) heterogeneity, and regulatory mechanisms that control molecular influx. Understanding these distinct permeability landscapes is crucial for developing novel antimicrobial strategies that can circumvent this fundamental resistance mechanism in high-priority pathogens.
The Gram-negative outer membrane is an asymmetric bilayer that performs the critical dual function of providing protection while permitting selective exchange of materials. The outer leaflet is composed primarily of lipopolysaccharide (LPS), while the inner leaflet contains phospholipids [10]. This unique architecture, particularly the densely packed LPS molecules with their strong lateral interactions and low fluidity, creates a formidable permeability barrier [10]. Permeation across this barrier occurs through two primary pathways: (1) lipid-mediated diffusion for hydrophobic antibiotics, and (2) porin-mediated general diffusion for hydrophilic antibiotics [10] [4]. The composition and regulation of these elements vary substantially between bacterial species, resulting in significant differences in intrinsic antibiotic susceptibility and resistance development mechanisms.
Structural and Functional Basis of OM Permeability
Lipopolysaccharide (LPS) Architecture and Barrier Function
The LPS layer provides a critical defensive barrier through its structural organization and physicochemical properties. A typical LPS molecule consists of three domains: lipid A (a glucosamine-based phospholipid), a core oligosaccharide, and a distal O-antigen polysaccharide [10]. The barrier function is significantly influenced by LPS composition:
- Lipid A structure: Typically contains six saturated fatty acid chains (compared to two in typical phospholipids), creating strong lateral interactions and low membrane fluidity [10]
- Core oligosaccharide: "Deep rough" mutants with truncated cores show dramatically increased sensitivity to hydrophobic antibiotics and detergents [10]
- Cationic bridges: Divalent cations (Mg²⁺, Ca²⁺) cross-bridge adjacent LPS molecules via their phosphate groups; disruption of these interactions by chelators (EDTA) or cationic antibiotics (polymyxins) compromises barrier integrity [10] [26]
Modifications to LPS structure, particularly through the addition of 4-aminoarabinose or phosphoethanolamine to lipid A phosphates, reduce the net negative charge of LPS, leading to a more tightly packed LPS layer and increased resistance to cationic antimicrobial peptides [10].
Porin Diversity and Permeation Pathways
Porins form water-filled channels that facilitate the passive diffusion of small hydrophilic molecules across the OM. The major porins differ significantly across the three target pathogens, as summarized in Table 1.
Table 1: Major Porin Profiles and Characteristics Across Target Pathogens
| Organism | Major Porins | Structural Features | Permeability Characteristics | Regulatory Influences |
|---|---|---|---|---|
| E. coli | OmpF, OmpC, OmpG, PhoE | Trimeric, general diffusion pores | Higher intrinsic permeability to hydrophilic compounds | Periplasmic H⁺ and K⁺ concentrations [24] |
| P. aeruginosa | OprF, Occ family (OpdK, OpdP, etc.) | OprF exists in open/closed conformations; Occ family shows substrate specificity | Naturally low permeability; restricted porin conductance [92] [26] | Unknown regulatory mechanisms |
| A. baumannii | OccAB1-OccAB5, Omp25, CarO, OprD | OccAB1 has largest pore; CarO for carbapenems | OccAB1 permits highest small-molecule uptake [93] | Limited information available |
The permeability differences are striking. P. aeruginosa exhibits naturally low OM permeability, with studies demonstrating that wild-type strains have significantly reduced porin-mediated uptake compared to antibiotic-supersusceptible mutants [92]. A. baumannii possesses five Occ (OM carboxylate channel) proteins with varying substrate specificities, with OccAB1 identified as having the largest pore and corresponding highest rates of small-molecule uptake [93].
Methodologies for Assessing OM Permeability
Fluorescence-Based Permeability Assays
Contemporary methods for evaluating OM permeability employ fluorescent probes that provide real-time, quantitative measurements of compound influx. The protocol described by Ma et al. (2021) offers a simultaneous assessment of both outer and inner membrane permeability [94]:
Key Reagents:
- N-phenyl-1-naphthylamine (NPN): Hydrophobic fluorophore that fluoresces upon intercalation into the OM; increased fluorescence indicates OM disruption
- Propidium iodide (PI): DNA-binding fluorophore excluded by intact membranes; penetration indicates compromised inner membrane integrity
Optimized Protocol:
- Culture bacteria under conditions identical to antibacterial assays (maintains physiological relevance)
- Harvest mid-log phase cells and wash with appropriate buffer
- Incubate with antibacterial molecules of interest under culture conditions
- Transfer aliquots to buffer containing fluorescent probes (avoiding interference from broth components)
- Measure fluorescence immediately using appropriate excitation/emission wavelengths (NPN: λex=350 nm, λem=420 nm; PI: λex=535 nm, λem=617 nm)
- Calculate permeability rates relative to untreated controls
This methodology provides a rapid, cost-effective assessment of membrane-permeabilizing effects that correlates well with results from minimum inhibitory concentration (MIC) and time-kill assays [94].
Single-Cell Imaging and Ionic Regulation Studies
Advanced imaging techniques have revealed dynamic regulation of porin permeability in response to metabolic changes. Zahn et al. (2019) employed single-cell imaging with fluorescent probes to demonstrate that porin permeability in E. coli is controlled by changes in periplasmic H⁺ and K⁺ concentrations [24]:
Experimental Workflow:
- Engineer bacterial strains with genetically encoded fluorescence sensors for periplasmic pH (pHuji) and K⁺ (GINKO2)
- Monitor ion concentration fluctuations in individual bacteria using microfluidic perfusion systems
- Measure porin permeability simultaneously using fluorescent substrates (2NBDG, Bocillin FL)
- Modulate ion gradients optogenetically (e.g., using light-activated proton pump ArchT)
- Correlate membrane potential (measured with QuasAr2) with porin permeability
This approach demonstrated that porin permeability increases during starvation (low periplasmic H⁺) to enhance nutrient uptake but decreases during periplasmic acidification to limit proton loss [24]. This metabolic control of porin permeability directly impacts antibiotic susceptibility patterns.
Diagram: Experimental Workflow for Single-Cell Analysis of Porin Permeability
Comparative Analysis of OM Permeability Mechanisms
Species-Specific Permeability Barriers
The three target pathogens employ distinct OM permeability strategies that reflect their environmental niches and intrinsic resistance profiles:
E. coli exhibits relatively higher OM permeability due to its abundant general diffusion porins (OmpF, OmpC). These trimeric porins allow passage of hydrophilic antibiotics up to ~600 Da [10] [4]. Regulation occurs through transcriptional control and post-translational modification in response to environmental conditions.
P. aeruginosa demonstrates exceptionally low OM permeability, a key factor in its intrinsic multidrug resistance. This results from several factors: (1) fewer porins per cell, (2) smaller porin diameter, and (3) preference for substrate-specific rather than general diffusion porins [92] [95]. The major porin OprF exists predominantly in a closed conformation with low permeability, with only a small fraction forming open channels [10].
A. baumannii possesses a diverse porin repertoire with the Occ protein family playing a major role in small-molecule passage [93]. Structural studies of OccAB1-OccAB4 reveal variations in channel dimensions that directly impact antibiotic permeability. Notably, modifications in porin expression patterns frequently contribute to acquired resistance in clinical isolates [93] [96].
Coordinated Regulation with Efflux Systems
OM permeability functions synergistically with efflux systems to limit intracellular antibiotic accumulation. The Resistance-Nodulation-Division (RND) family of efflux pumps, present exclusively in Gram-negative bacteria, work in concert with the OM barrier to create a highly effective defense system [26]. This coordination is particularly well-developed in P. aeruginosa, which expresses multiple RND systems that can export a broad spectrum of antibiotics [95] [26]. Modifications in OM permeability often coincide with increased efflux pump expression, creating a compounded resistance phenotype that is difficult to overcome.
Research Reagent Solutions for OM Permeability Studies
Table 2: Essential Research Reagents for OM Permeability Investigations
| Reagent/Category | Specific Examples | Research Application | Key Considerations |
|---|---|---|---|
| Fluorescent Probes | NPN, Propidium iodide, 2NBDG, Hoechst, Bocillin FL | Quantitative permeability assessment; real-time influx measurement | Select probes based on hydrophilicity/hydrophobicity match to antibiotics of interest |
| Ionophores & Modulators | CCCP (protonophore), Valinomycin (K⁺ ionophore), Polymyxin B nonapeptide | Dissect ionic influences on porin conductance; membrane perturbation studies | Use sub-inhibitory concentrations to avoid collateral effects on viability |
| Genetic Tools | Fluorescent protein sensors (pHuji, GINKO1/2, QuasAr2), Optogenetic actuators (ArchT) | Monitor ion fluxes; manipulate membrane potential in real-time | Requires specialized bacterial strains; microfluidic systems enhance temporal resolution |
| Permeabilizers | EDTA, Tris, Polymyxin B derivatives | Positive controls for barrier disruption; study LPS-cation interactions | Concentration-dependent effects; species-specific sensitivity patterns |
| Porin-Specific Tools | Purified porins, Liposome swelling assays, Planar lipid bilayers | Define channel characteristics; measure conductance parameters | Technical expertise required; provides mechanistic versus cellular insights |
Implications for Antibiotic Resistance and Therapeutic Development
The species-specific differences in OM permeability have direct consequences for antibiotic resistance and drug development:
E. coli serves as a model for understanding porin-mediated resistance, where mutations altering porin expression or structure (e.g., OmpF/C) reduce antibiotic influx, particularly when combined with periplasmic β-lactamases [4]. Recent evidence indicates metabolic regulation of porin permeability via periplasmic H⁺ and K⁺ concentrations, explaining increased ciprofloxacin resistance during growth on lipids [24].
P. aeruginosa's impermeable OM synergizes with its extensive efflux systems and enzymatic degradation mechanisms to create formidable multidrug resistance [95]. This combination presents a significant challenge for antibiotic design, requiring compounds that can either penetrate the OM barrier or bypass it entirely.
A. baumannii demonstrates how porin alterations contribute to carbapenem resistance, particularly through changes in CarO and OprD-like channels [93] [96]. Recent work reveals that colistin resistance via mcr genes damages OM integrity, creating a fitness cost that increases susceptibility to hydrophobic antibiotics like rifampicin [96].
Diagram: Interplay Between OM Permeability and Antibiotic Resistance Mechanisms
Future Perspectives and Concluding Remarks
The comparative analysis of OM permeability across E. coli, P. aeruginosa, and A. baumannii reveals both shared principles and distinct adaptations that contribute to antibiotic resistance. Future research should focus on:
- Structural characterization of less-studied porins from P. aeruginosa and A. baumannii to identify potential passageways for antimicrobial compounds
- Metabolic regulation of porin expression and conductance as a therapeutic target to enhance antibiotic penetration
- Synergistic combinations that exploit fitness costs associated with membrane modifications, such as colistin with hydrophobic antibiotics against resistant strains [96]
- Computational approaches to model molecular transit through species-specific porin channels for rational antibiotic design
Understanding the nuanced differences in OM permeability mechanisms among these clinically important pathogens provides a foundation for developing novel strategies to overcome this fundamental antibiotic resistance barrier. The continued development of sophisticated research tools, particularly those enabling real-time, single-cell analysis of permeability dynamics, will be essential for these efforts.
The intrinsic and acquired resistance of Pseudomonas aeruginosa to a wide range of antibiotics represents a critical challenge in clinical settings worldwide. A major factor contributing to this resilience is the low permeability of its outer membrane, which functions as a formidable barrier to antimicrobial agents [97]. Unlike other Gram-negative bacteria such as Escherichia coli, P. aeruginosa lacks classic general porins and instead possesses predominantly narrow, substrate-specific channels [98]. Among these, the OprD porin family plays a pivotal role in nutrient uptake and antibiotic susceptibility. OprD, in particular, has garnered significant research attention due to its central function in facilitating the influx of basic amino acids and carbapenem antibiotics [99]. The loss or impaired function of OprD is a predominant mechanism conferring resistance to carbapenems, especially imipenem, making it a key research focus in understanding and combating multidrug-resistant P. aeruginosa infections [99] [100]. This review delves into the structure, function, and regulation of OprD, frames its role within the broader context of outer membrane permeability and antibiotic resistance, and summarizes contemporary experimental approaches for its study.
OprD: Structure, Function, and Mechanism of Antibiotic Permeation
Historical Identification and Genetic Locus
The identification of OprD is intrinsically linked to the emergence of imipenem resistance. Resistance to imipenem was identified concurrently with or even before the drug's clinical approval in the USA in 1987 [99]. Early studies consistently correlated this resistance phenotype with the absence of a 45-49 kilodalton (kDa) outer membrane protein, later identified as OprD [99]. The gene encoding OprD, oprD, is located between 71 and 75 minutes on the P. aeruginosa PAO1 chromosome and consists of 1332 base pairs [99].
Protein Topology and Channel Architecture
OprD belongs to the porin family, sharing 41% to 58% sequence similarity with other members, and is predicted to adopt a typical porin topology [99]. This structure comprises a 16-strand transmembrane β-barrel traversing the outer membrane. The barrel is formed by seven short turn sequences on the periplasmic side that act as hinges, connecting eight loop (L) structures exposed to the external environment [99]. The channel formed by OprD is notably narrower than general porins like OmpF in E. coli, which is a fundamental reason for the characteristically low outer membrane permeability of P. aeruginosa [99].
Table 1: Key Loops in OprD Porin and Their Functional Roles
| Loop Identifier | Functional Role | Impact of Deletion/Mutation |
|---|---|---|
| Loop 2 | Primary binding site for imipenem; entrance for basic amino acids [99]. | Deletion induces partial resistance to imipenem and meropenem; eliminates imipenem-blocked KCl conductance [99]. |
| Loop 3 | Serves as a passage channel for imipenem [99]. | Deletion results in failure to reconstitute imipenem susceptibility [99]. |
| Loops 1, 5, 6, 7, 8 | Not directly involved in imipenem passage [99]. | Deletion does not alter imipenem susceptibility [99]. Mutations in Loop 7 can increase meropenem susceptibility [99]. |
Molecular Basis of Substrate Specificity and Carbapenem Uptake
OprD functions as a specific channel for the uptake of basic amino acids and small peptides, such as arginine [99] [101]. The structural similarity between the side chain of imipenem and basic amino acids like arginine allows the antibiotic to utilize OprD for entry into the cell [101]. Functional studies have pinpointed loops 2 and 3 as the critical structural determinants for this process. Loop 2 contains the direct binding site for imipenem, while loop 3 facilitates the passage of the molecule through the channel [99]. Any mutation, substitution, or deletion within these loops that alters the channel conformation can lead to carbapenem resistance by hindering antibiotic uptake [99]. Furthermore, the presence of natural substrates like amino acids can compete with carbapenems for passage through OprD, potentially modulating antibiotic efficacy in vivo [99].
Figure 1: Schematic of Substrate and Antibiotic Permeation through the OprD Porin. Loop 2 and Loop 3 are critical for the uptake of carbapenems like imipenem, which competes with natural substrates such as basic amino acids for passage through the narrow channel.
OprD-Mediated Resistance and Regulatory Networks
OprD Deficiency as a Primary Resistance Mechanism
The loss or functional impairment of OprD is the most prevalent mechanism for carbapenem resistance in P. aeruginosa [99]. This deficiency confers a basal level of resistance, particularly to imipenem [99]. A recent study on clinical isolates that were resistant only to carbapenems (CROPA) provided compelling evidence, directly linking this resistance profile to mutations and loss of OprD function, without the involvement of other major mechanisms like carbapenemase production or efflux pump overexpression [100]. The reintroduction of a functional oprD gene in these strains restored carbapenem susceptibility, confirming OprD's central role [100].
Table 2: Mechanisms of OprD Inactivation and Their Consequences in P. aeruginosa
| Mechanism of Inactivation | Molecular Consequence | Phenotypic Outcome | Citation |
|---|---|---|---|
| Amino Acid Mutations | Conformational changes in loops 2 and 3, impairing imipenem binding and passage [99]. | Carbapenem resistance (often imipenem-specific) [99]. | |
| Premature Termination | Truncated, non-functional OprD protein [100]. | Loss of carbapenem uptake [100]. | |
| Insertion Elements (e.g., IS256) | Disruption of the oprD coding sequence [100]. | Abrogated OprD expression and carbapenem resistance [100]. | |
| Transcriptional Regulation | Downregulation of oprD gene expression [99]. | Reduced porin levels and increased MICs [99]. |
Complex Regulation of OprD Expression
OprD expression is highly regulated at both transcriptional and post-transcriptional levels by a suite of environmental and cellular factors [99]. This complex regulation underscores the adaptability of P. aeruginosa.
- Metals and Bioactive Molecules: The presence of certain metals and small bioactive molecules can repress oprD transcription [99].
- Amino Acid Availability: As OprD is a nutrient uptake channel, its expression is modulated by the availability of its substrates, such as basic amino acids [99].
- Efflux Pump Regulators: The expression of OprD is intricately linked with the regulation of efflux pumps. Notably, the overexpression of the MexAB-OprM efflux system is often accompanied by the downregulation of OprD, leading to a synergistic increase in resistance levels [99]. This combination can render the bacterium highly resistant not only to imipenem but also to meropenem and doripenem [99].
- Growth Phase-Dependent Expression: Recent evidence indicates that the expression of porins, including OprD and its homolog OpdP, varies throughout the bacterial growth cycle. OpdP expression increases as the culture enters the stationary phase, inversely correlating with OprD expression, suggesting a sophisticated, growth-phase-dependent permeability program [101].
The OprD Porin Family and the Occ Subgroups
OprD is the namesake of a larger family of substrate-specific porins in P. aeruginosa, known as the outer membrane carboxylate channel (Occ) family [101]. This family is divided into the OprD (OccD) and OpdK (OccK) subfamilies, collectively comprising 18 homologs [101]. These porins exhibit high substrate specificity. For instance:
- OprP is highly selective for phosphate ions [98].
- OprO, which shares 74% sequence identity with OprP, is selective for pyrophosphate (diphosphate) [98].
- OpdP (OccD3), sharing 51% sequence identity with OprD, is associated with glycine-glutamate dipeptide translocation and has also been implicated in meropenem uptake [101].
The specificity of these highly similar channels, such as OprP and OprO, is determined by a small number of residues in the central constriction region. For example, substituting two key residues in OprO (F62Y and D114Y) can interchange its pyrophosphate specificity to the phosphate specificity characteristic of OprP [98]. This highlights the potential for rational design of channel properties.
Experimental Methodologies for Studying OprD Function and Expression
Key Experimental Workflow
A multidisciplinary approach is required to fully characterize OprD-mediated resistance in clinical and laboratory isolates. The following workflow integrates genotypic, phenotypic, and functional assays.
Figure 2: Experimental Workflow for Characterizing OprD-Mediated Resistance. A comprehensive analysis integrates genotypic data from sequencing with phenotypic susceptibility testing and functional validation.
Detailed Methodologies
Antimicrobial Susceptibility Testing (AST)
Purpose: To determine the Minimum Inhibitory Concentration (MIC) of carbapenems and other antibiotics, establishing the resistance phenotype. Protocol:
- Strain Preparation: Cultivate purified P. aeruginosa isolates in Mueller-Hinton (MH) broth for 24 hours [100].
- Automated Testing: Use systems like the VITEK 2 COMPACT with Gram-negative susceptibility cards (e.g., N335) according to manufacturer instructions [100].
- Interpretation: Interpret results based on current Clinical and Laboratory Standards Institute (CLSI) guidelines (e.g., M100) [100]. Control strains like P. aeruginosa ATCC 27853 must be included for quality assurance. Output: MIC values for imipenem, meropenem, and other anti-pseudomonal drugs, categorizing isolates as sensitive, intermediate, or resistant.
Genetic Analysis of OprD
Purpose: To identify mutations, insertions, or deletions in the oprD gene that explain the resistance phenotype. Protocol:
- DNA Extraction: Grow bacteria in LB medium to an OD600 of ~0.8. Extract genomic DNA using commercial kits or a boiling method [100].
- Whole-Genome Sequencing (WGS): Prepare libraries using a kit (e.g., Illumina TruSeqTM Nano DNA Sample Prep Kit) and sequence on a platform such as Illumina NovaSeq6000 [100].
- Bioinformatic Analysis:
- Assembly: Assemble raw sequences using software like ABySS v2.0.2 and polish with GapCloser v1.12 [100].
- Annotation: Annotate predicted genes using BLAST against databases (NR, Swiss-Prot, etc.) [100].
- Variant Calling: Map sequences to a reference genome (e.g., PAO1) and identify variants in the oprD gene. Also screen for known carbapenemase genes (e.g., VIM, IMP, NDM) via PCR [100]. Output: A comprehensive report on oprD mutations (e.g., premature stop codons, IS element insertions, missense mutations in critical loops) and the presence/absence of carbapenemases.
Expression Analysis by RT-qPCR and SDS-PAGE
Purpose: To correlate genetic findings with the expression level of OprD at the mRNA and protein level. Protocol:
- RT-qPCR for mRNA Level:
- RNA Extraction: Use a commercial kit (e.g., MiniBEST Universal RNA Extraction Kit) to isolate total RNA [100].
- cDNA Synthesis: Reverse transcribe RNA to cDNA using a reagent kit (e.g., PrimeScript RT reagent Kit) [100].
- qPCR Amplification: Perform qPCR using a mix (e.g., 2× SYBR Green qPCR Mix) and gene-specific primers for oprD. Normalize transcript levels using a housekeeping gene (e.g., rpsL). A reduction of ≥50% in oprD expression compared to a reference strain (e.g., PAO1) is considered significant [100].
- SDS-PAGE for Protein Level:
- Membrane Protein Preparation: Solubilize outer membrane proteins from bacterial cultures.
- Electrophoresis: Separate proteins via SDS-PAGE and visualize using Coomassie blue staining or perform Western immunoblotting for specific detection of OprD [100]. Output: Quantitative data on oprD transcript levels and qualitative/quantitative assessment of OprD protein presence.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents and Materials for OprD Research
| Reagent / Material | Specific Example | Function in Research |
|---|---|---|
| Reference Strains | P. aeruginosa PAO1 (wild-type), ATCC 27853 (quality control) [100]. | Essential controls for genetic and phenotypic comparisons; used for AST quality assurance. |
| Culture Media | Mueller-Hinton (MH) Broth/Agar, Luria-Bertani (LB) Broth [100]. | Standardized media for antimicrobial susceptibility testing and general bacterial cultivation. |
| Antibiotic Susceptibility Test System | VITEK 2 COMPACT with N335 cards, Broth microdilution panels [100]. | Determines Minimum Inhibitory Concentrations (MICs) for carbapenems and other antibiotics. |
| DNA/RNA Extraction Kits | MiniBEST Universal RNA Extraction Kit, various genomic DNA extraction kits [100]. | High-quality nucleic acid isolation for downstream genomic and transcriptomic analyses. |
| Sequencing & Cloning Tools | Illumina TruSeqTM Nano DNA Sample Prep Kit, expression plasmids (e.g., pB22 for OprO) [98] [100]. | Whole-genome sequencing library prep and molecular cloning for functional validation studies. |
| qPCR Reagents | PrimeScript RT reagent Kit, 2× SYBR Green qPCR Mix [100]. | Quantitative analysis of gene expression (e.g., oprD, efflux pump genes). |
OprD represents a critical linchpin in the complex interplay between outer membrane permeability and antibiotic resistance in P. aeruginosa. Its role as a specific gateway for carbapenems makes it a high-value research target. The evolving understanding of its regulation, its position within the extensive Occ porin family, and its interaction with other resistance mechanisms like efflux pumps, provides a more complete picture of bacterial adaptability. Future research directions should focus on leveraging high-resolution structural data to inform the design of novel antibiotics that can bypass OprD-dependent pathways or exploit its structure for enhanced uptake. Furthermore, a deeper investigation into the regulatory networks that control OprD expression could reveal targets for adjuvants that potentiate the activity of existing carbapenems. As the threat of carbapenem-resistant P. aeruginosa continues to grow, a refined understanding of OprD will be indispensable for guiding clinical diagnostics and developing the next generation of therapeutic strategies.
The outer membrane (OM) of Gram-negative bacteria serves as a formidable permeability barrier, contributing significantly to intrinsic antibiotic resistance. For the nosocomial pathogen Acinetobacter baumannii, the structure of lipooligosaccharide (LOS), particularly the lipid A domain, is a critical determinant of OM integrity. Unlike many Gram-negative pathogens that synthesize hexa-acylated lipid A, A. baumannii produces a predominant hepta-acylated lipid A species under standard growth conditions via a unique, PagP-independent biosynthetic pathway [102]. This structural modification profoundly influences the biophysical properties of the OM, reducing permeability to many antimicrobials and contributing to the pathogen's resilience in hospital environments. This review examines the biosynthesis, structural characteristics, and functional consequences of hepta-acylated lipid A in A. baumannii, with emphasis on its role in antibiotic resistance and future therapeutic targeting.
Biosynthesis and Structural Features of A. baumannii Lipid A
Acyltransferase Machinery
The biosynthesis of hepta-acylated lipid A in A. baumannii involves specialized acyltransferases that diverge from the canonical Raetz pathway:
- LpxLAb (A. baumannii LpxL): Transfers the first lauroyl (C12:0) acyl chain to the precursor Kdo2-lipid IVA [102]
- LpxMAb (A. baumannii LpxM): Functions as a dual acyltransferase, transferring two lauroyl groups onto lipid A to generate the characteristic hepta-acylated structure [102]
This biosynthetic pathway differs fundamentally from those in Escherichia coli and Salmonella, where LpxL and LpxM add one laurate and one myristate, respectively, producing only hexa-acylated lipid A [102]. The hepta-acylated lipid A produced by this pathway represents the most abundant glycolipid species on the surface of A. baumannii [103].
Additional Structural Modifications
Beyond the core hepta-acylated structure, A. baumannii lipid A undergoes further modifications that enhance its protective functions:
- 2-Hydroxylation: The enzyme LpxO catalyzes the 2-hydroxylation of the laurate transferred by LpxLAb, producing 2-hydroxylaurate modifications that provide additional protection against cationic antimicrobial peptides (CAMPs) like polymyxin B and human β-defensin 3 [104]
- Phosphoethanolamine and Galactosamine Addition: These modifications, regulated by the PmrAB two-component system, are implicated in colistin resistance [104]
Table 1: Key Enzymes in A. baumannii Lipid A Biosynthesis and Modification
| Enzyme | Function | Effect on Lipid A Structure | Phenotypic Consequence |
|---|---|---|---|
| LpxLAb | Transfers first lauroyl (C12:0) acyl chain | Adds secondary acyl chain at 2' position | Essential for hepta-acylation pathway [102] |
| LpxMAb | Dual acyltransferase transferring two lauroyl groups | Generates hepta-acylated lipid A species | Confers resistance to CAMPs and promotes desiccation survival [102] |
| LpxO | Catalyzes 2-hydroxylation of laurate | Adds hydroxyl group to secondary acyl chain | Enhances resistance to polymyxins and human defensins [104] |
| PmrC/EptA | Adds phosphoethanolamine | Modifies phosphate groups | Contributes to colistin resistance [104] |
| NaxD | Adds galactosamine | Modifies phosphate groups | Mediates colistin resistance [104] |
Biophysical Consequences on Outer Membrane Packing
The structural characteristics of hepta-acylated lipid A significantly enhance the packing density of the outer membrane, creating a formidable barrier to antimicrobial penetration.
Membrane Organization and Stability
The additional acyl chain in hepta-acylated lipid A improves the hydrophobic interactions between adjacent LOS molecules:
- Enhanced Packing Density: The extra lauroyl chain allows for tighter packing of lipid A molecules in the outer leaflet of the OM, reducing void spaces and restricting passive diffusion of small molecules [59]
- Improved Membrane Ordering: Molecular dynamics simulations of various lipid A structures indicate that increased acyl chain number correlates with larger area per lipid and enhanced hydrophobic thickness [105]
- Barrier Fortification: The hepta-acylated structure reinforces the LPS portion of the OM barrier, making it less permeable to both hydrophobic and hydrophilic compounds [102]
Role in Desiccation Survival
The enhanced membrane packing conferred by hepta-acylated lipid A has functional implications beyond antibiotic resistance:
- LpxMAb Essentiality: A. baumannii mutants lacking LpxMAb show dramatically reduced survival under desiccative conditions, highlighting the importance of hepta-acylation for persistence on hospital surfaces [102]
- Membrane Integrity: The tightly packed OM prevents excessive water loss and maintains membrane integrity during extended dry periods [102]
Impact on Outer Membrane Permeability and Antibiotic Resistance
Permeability to Antibiotics
The fortified OM of A. baumannii presents a significant challenge for antibiotic penetration:
- Porin-Dependent Permeation: A. baumannii lacks classical porins and instead utilizes substrate-specific porins with narrower channels. Among these, OccAB1 has the largest pore and represents the most promising route for small-molecule antibiotic uptake [106] [59]
- Restricted Access: The tight packing of hepta-acylated lipid A reduces the permeability of charged and hydrophilic molecules, forcing antibiotics to rely on specific porin pathways for cellular entry [59]
- Amphiphilic Compound Permeation: Interestingly, the OM becomes more permeable to amphiphilic compounds due to the altered membrane properties conferred by hepta-acylation [59]
Resistance to Cationic Antimicrobial Peptides
Hepta-acylated lipid A provides enhanced resistance to host defense mechanisms and last-resort antibiotics:
- Reduced Negative Charge: The additional acyl chain may partially shield the negative charges of the lipid A phosphate groups, reducing electrostatic attraction to cationic peptides [102]
- Steric Hindrance: The bulkier lipid A headgroup creates steric hindrance that limits access of CAMPs to their membrane targets [102] [104]
- Polymyxin Resistance: Hepta-acylated lipid A promotes resistance to polymyxin antibiotics (including colistin), which are often last-resort treatments for multidrug-resistant A. baumannii infections [102]
- Synergistic Effects: The combination of hepta-acylation and 2-hydroxylation provides maximal protection against diverse CAMPs [104]
Table 2: Antibiotic Resistance Profiles Associated with A. baumannii Lipid A Modifications
| Lipid A Modification | Resistance Profile | Proposed Mechanism | Clinical Impact |
|---|---|---|---|
| Hepta-acylation (LpxMAb-dependent) | Resistance to vertebrate CAMPs and polymyxins [102] | Enhanced OM packing, reduced membrane fluidity | Compromises efficacy of last-resort antibiotics [102] |
| 2-Hydroxylation (LpxO-dependent) | Enhanced resistance to polymyxin B, colistin, and human β-defensin 3 [104] | Altered CAMP binding affinity through hydroxylation | Promotes survival in human whole blood and invertebrate models [104] |
| Phosphoethanolamine addition (PmrC-dependent) | Colistin resistance [104] | Neutralization of lipid A negative charges | Associated with mutations in PmrAB two-component system [104] |
| Complete LOS loss (lpxA/C/D mutations) | Colistin resistance [104] | Elimination of colistin target | Dramatic reduction in virulence and fitness [104] |
Experimental Approaches for Studying Lipid A Structure-Function Relationships
Genetic Manipulation of Lipid A Biosynthesis
The functional characterization of hepta-acylated lipid A relies on well-established genetic techniques:
- Gene Knockout Strategies: Construction of isogenic mutants in lpxLAb, lpxMAb, and lpxO using homologous recombination or recombineering systems to elucidate the contribution of each enzyme to lipid A biosynthesis and modification [102] [104]
- Complementation Studies: Restoration of wild-type genes in mutant backgrounds to confirm genotype-phenotype relationships and rule out polar effects [104]
- Conditional Expression Systems: Regulated expression of lipid A modification enzymes to assess the dynamic adaptation of the OM under different environmental conditions
Structural Analysis of Lipid A Species
Detailed structural characterization is essential for understanding the relationship between lipid A chemistry and OM function:
- Lipid A Extraction: Isolation of lipid A from whole cells using mild acid hydrolysis or an ammonium hydroxide-isobutyric acid method [104]
- Mass Spectrometry Analysis: Structural characterization using MALDI-TOF mass spectrometry in negative-ion mode to determine molecular mass and acylation patterns [104]
- Thin-Layer Chromatography: Assessment of global membrane lipid composition to ensure specific changes in lipid A without broader disruption of other membrane lipids [104]
Functional Assays for Membrane Properties
The functional consequences of lipid A modifications can be quantified through various biochemical and biophysical assays:
- Membrane Permeability Assays: Evaluation of OM integrity using hydrophobic fluorescent probes such as 1-N-phenylnaphthylamine (NPN), which exhibits increased fluorescence when partitioning into disrupted OMs [104]
- Antimicrobial Susceptibility Testing: Determination of minimum inhibitory concentrations (MICs) against CAMPs, polymyxins, and conventional antibiotics using standard broth microdilution methods [96]
- Surface Stress Sensitivity: Assessment of bacterial susceptibility to detergents (e.g., SDS) and chelators (e.g., EDTA) to evaluate OM stability [96]
Diagram 1: Biosynthetic pathway of hepta-acylated lipid A in A. baumannii and its functional consequences on outer membrane properties and antibiotic resistance.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Studying A. baumannii Lipid A and OM Permeability
| Reagent/Technique | Specific Example | Research Application | Technical Considerations |
|---|---|---|---|
| Genetic tools | A. baumannii recombineering system [102] | Construction of lipid A biosynthetic mutants | Essential for generating isogenic mutants in lipid A pathway |
| Mass spectrometry | MALDI-TOF MS [104] | Structural characterization of lipid A species | Requires lipid A extraction and purification; negative-ion mode for lipid A |
| Membrane probes | 1-N-phenylnaphthylamine (NPN) [104] | Assessment of outer membrane integrity | Fluorescence increases with OM disruption |
| Antimicrobial assays | Broth microdilution for MIC determination [96] | Evaluation of susceptibility to CAMPs and antibiotics | Follow CLSI guidelines for standardization |
| Stress sensitivity assays | SDS and EDTA exposure [96] | Assessment of OM stability | Serial dilution spotting for qualitative assessment |
| Whole blood survival | Human whole blood model [104] | Evaluation of serum resistance | Measures complement sensitivity and phagocytosis resistance |
| Invertebrate models | Galleria mellonella [104] | Assessment of in vivo virulence | Correlates with mammalian models for initial screening |
The hepta-acylated lipid A of A. baumannii represents a masterful adaptation that enhances OM packing and reduces permeability to antimicrobials. This structural modification contributes significantly to the pathogen's environmental resilience and resistance to last-resort antibiotics. Future therapeutic strategies may include:
- LpxMAb Inhibitors: Development of small molecules targeting the unique dual acyltransferase activity of LpxMAb to disrupt hepta-acylated lipid A biosynthesis [102]
- Permeability Enhancers: Compounds that specifically disrupt the tight packing of hepta-acylated lipid A to increase antibiotic penetration [59]
- Combination Therapies: Leveraging the increased susceptibility of colistin-resistant strains with damaged OM to macromolecular antibiotics like rifampicin [96]
Understanding the structure-function relationships of A. baumannii lipid A provides critical insights for developing novel therapeutic approaches against this formidable pathogen.
The intrinsic resistance of Gram-negative bacteria to many antibiotics is largely attributable to the formidable barrier presented by their cell envelope [10] [26]. This complex structure, comprising an outer membrane (OM), an inner membrane (IM), and a periplasmic space in between, sharply limits the intracellular accumulation of antibacterial compounds, rendering many potential therapeutics ineffective [4] [107]. A critical step in overcoming this challenge and developing novel antibiotics is the accurate quantification of a compound's ability to traverse these layers and reach its intracellular target [107]. This guide provides an in-depth review of contemporary biochemical, biophysical, and microbiological assays essential for validating compound penetration into Gram-negative bacteria, a core aspect of research focused on defeating outer membrane-mediated antibiotic resistance.
The Gram-Negative Barrier: Structure and Permeability Pathways
The Gram-negative outer membrane is an asymmetric bilayer. Its inner leaflet consists of phospholipids, while the outer leaflet is composed primarily of lipopolysaccharides (LPS) [10] [4]. LPS molecules are densely packed, their negative charges stabilized by divalent cations (Mg²⁺, Ca²⁺), creating a highly impermeable barrier to hydrophobic molecules [10] [26]. Embedded within this membrane are porins, β-barrel proteins that form water-filled channels permitting the passive diffusion of small, hydrophilic molecules [10] [108].
Compound penetration occurs primarily via two pathways:
- Porin-Mediated Diffusion: This is the main route for hydrophilic antibiotics, such as β-lactams. Porins like OmpF and OmpC in E. coli act as molecular sieves, typically excluding molecules above ~600 Da [109] [110].
- Lipid-Mediated Diffusion: Hydrophobic antibiotics, including macrolides and rifamycins, diffuse directly through the lipid bilayer of the outer membrane [10] [4]. The tight packing of LPS chains makes this pathway inherently inefficient for many compounds.
Understanding which pathway a compound utilizes is fundamental to optimizing its design and interpreting accumulation assays. Furthermore, the synergy between this low-permeability barrier and active efflux pumps, which eject compounds back into the extracellular environment, dramatically reduces intracellular antibiotic concentrations [26] [107].
Quantitative Whole-Cell Accumulation Assays
Mass Spectrometry-Based Methods
Mass spectrometry (MS) offers a direct and label-free approach to quantify a compound's uptake into bacterial cells. These methods can be broadly categorized into two workflows [107]:
- Intracellular Concentration Measurement: Cells are exposed to the antibiotic, then rapidly separated from the medium (e.g., via centrifugation through silicone oil). The cell pellet is lysed, and the intracellular antibiotic concentration is quantified using LC-MS/MS. This method, while labor-intensive, provides absolute values of compound accumulation [107].
- Extracellular Concentration Depletion: This approach indirectly measures uptake by monitoring the decrease in antibiotic concentration from the surrounding medium over time. It is less disruptive and allows for continuous monitoring, though it may be less sensitive for compounds that accumulate modestly [107].
Experimental Protocol: Intracellular Accumulation via LC-MS/MS
- Step 1: Antibiotic Exposure. A concentrated bacterial culture is diluted into fresh, pre-warmed medium containing the antibiotic of interest at a desired concentration (e.g., 1-10x MIC). A control sample without antibiotic is also prepared.
- Step 2: Incubation & Uptake. The culture is incubated under standard growth conditions (e.g., 37°C with shaking) for a defined period to allow antibiotic accumulation.
- Step 3: Rapid Separation. Aliquots of the culture are centrifuged at high speed (e.g., 14,000 x g) through a dense, immiscible layer like silicone oil. This rapidly separates cells from the extracellular medium.
- Step 4: Cell Lysis and Extraction. The supernatant is carefully removed, and the cell pellet is lysed using a method such as boiling in SDS or bead beating in the presence of an appropriate solvent to extract the antibiotic.
- Step 5: LC-MS/MS Analysis. The lysate is clarified by centrifugation, and the supernatant is analyzed by LC-MS/MS. The antibiotic concentration is determined by comparing the signal to a standard curve. Data are normalized to total cellular protein or cell count.
Fluorescence-Based Assays and Single-Cell Microscopy
Fluorescence-based techniques provide powerful tools for visualizing and quantifying drug uptake at both population and single-cell levels.
- NPN Uptake Assay: The fluorescent probe 1-N-phenylnaphthylamine (NPN) is weakly fluorescent in aqueous environments but exhibits strong fluorescence upon entering the cell and binding to hydrophobic membrane interiors. This property is exploited to assay outer membrane permeability [109]. Treatment with a permeabilizing agent (e.g., the LolA inhibitor MAC13243 or polymyxin derivatives) causes a quantifiable increase in NPN fluorescence, indicating a compromised OM barrier [109].
Experimental Protocol: NPN Uptake to Assess OM Permeability
- Step 1: Cell Preparation. Grow bacteria to mid-log phase. Harvest cells by centrifugation and wash/resuspend in an appropriate buffer (e.g., HEPES or PBS).
- Step 2: Assay Setup. In a black-walled 96-well plate, mix bacterial suspension with NPN to a final concentration of ~10 µM.
- Step 3: Baseline Reading. Measure fluorescence (excitation ~355 nm, emission ~405 nm) to establish a baseline.
- Step 4: Compound Addition. Add the test compound (e.g., MAC13243 at a sub-inhibitory concentration) or a positive control (e.g., polymyxin B nonapeptide) to the well.
- Step 5: Fluorescence Monitoring. Immediately monitor fluorescence over time. An increase in fluorescence relative to an untreated control indicates increased outer membrane permeability.
Deep UV Microscopy: This label-free technique leverages the innate fluorescence of certain antibiotics (e.g., quinolones) when imaged under deep UV light. It allows for the quantification of antibiotic accumulation within single cells, revealing cell-to-cell heterogeneity in uptake that bulk methods might obscure [107].
Fluorescent Antibiotic Derivatives: Chemically modified antibiotics tagged with fluorophores (e.g., BODIPY-labeled vancomycin) enable direct visualization of their localization and accumulation via fluorescence microscopy [107]. This is particularly useful for antibiotics that target extracellular structures like the peptidoglycan in the periplasm.
Biophysical and Model System Assays
Electrophysiology in Black Lipid Membranes (BLM)
Black lipid membrane (BLM) experiments provide unparalleled, high-resolution insights into the molecular interaction of antibiotics with specific porin pathways [107].
Experimental Protocol: Single-Channel Recording with Porins
- Step 1: Membrane Formation. A lipid solution (e.g., diphytanoylphosphatidylcholine in decane) is painted across a small aperture (~100 µm diameter) separating two electrolyte-filled chambers (e.g., 1 M KCl). The lipid bilayer thins to a "black" membrane.
- Step 2: Porin Incorporation. Purified, detergent-solubilized porin (e.g., OmpF) is added to one chamber (the cis side). Individual porin molecules spontaneously insert into the bilayer.
- Step 3: Ion Current Measurement. A voltage (e.g., +50 mV to +100 mV) is applied across the membrane. The insertion of a single porin channel causes a discrete jump in ionic current. The conductance and stability of this open channel are recorded.
- Step 4: Antibiotic Addition. The antibiotic is added to the cis chamber. As antibiotic molecules diffuse through the porin channel, they transiently block the ion flow, causing characteristic rapid flickers or partial blockades in the current trace.
- Step 5: Data Analysis. The frequency and duration of these blockades are analyzed to determine the kinetics of antibiotic translocation (association and dissociation rates) and the affinity of the antibiotic-porin interaction.
Biomimetic Liposome Assays
Liposomes, or lipid vesicles, are synthetic systems used to model the bacterial membranes. Swelling assays, for instance, monitor the influx of solutes into liposomes reconstituted with porins by measuring the resulting increase in liposome volume (light scattering) [107]. More advanced assays use liposomes encapsulating a fluorescent dye; the influx of an antibiotic that quenches the dye's fluorescence can be monitored in real-time [107].
Table 1: Key Reagent Solutions for Penetration Assays
| Research Reagent | Function/Application in Assays | Key Considerations |
|---|---|---|
| Silicone Oil | Rapid separation of cells from medium during centrifugation for MS-based accumulation assays. | Density must be chosen to form a distinct layer between cells and supernatant. |
| 1-N-phenylnaphthylamine (NPN) | Fluorescent probe for assessing outer membrane integrity and permeability. | Fluorescence increases in hydrophobic environments; use fresh stock solutions. |
| Polymyxin B Nonapeptide (PMBN) | Positive control for OM permeabilization in NPN assays; disrupts LPS organization. | Lacks the bactericidal activity of full polymyxin B, making it ideal for permeabilization studies. |
| Purified Porins (e.g., OmpF, OmpC) | For reconstitution into Black Lipid Membranes (BLM) or liposomes to study specific translocation pathways. | Requires expression and purification from E. coli or other hosts; must be kept in detergent to prevent aggregation. |
| Diphytanoylphosphatidylcholine | Synthetic lipid used to form stable, high-resistance bilayers for BLM electrophysiology. | Forms long-lasting, fluid bilayers ideal for single-channel recording. |
Microbiological and Genetic Susceptibility Assays
While not direct measures of accumulation, microbiological assays provide a functional readout of a compound's ability to penetrate the cell envelope and reach its target.
Synergy Checkerboard Assay
This assay determines if the combination of a permeabilizing agent and a large-scaffold antibiotic results in synergistic activity [109].
Experimental Protocol
- Step 1: Plate Setup. Prepare a 96-well plate with a two-dimensional dilution series. One antibiotic (e.g., a large-scaffold drug like erythromycin) is diluted along the rows, while the second compound (e.g., a permeabilizer like MAC13243) is diluted down the columns.
- Step 2: Inoculation. Add a standardized inoculum of bacteria to each well.
- Step 3: Incubation and Growth Assessment. Incubate the plate and measure bacterial growth (e.g., by OD600) after a set time.
- Step 4: FICI Calculation. The Fractional Inhibitory Concentration Index (FICI) is calculated for each well where growth was inhibited. FICI = (MIC of drug A in combination / MIC of drug A alone) + (MIC of drug B in combination / MIC of drug B alone). A FICI of ≤ 0.5 is generally considered synergistic [109].
Genetic Constructions for Validation
Genetic tools allow researchers to directly link a specific gene to permeability.
- Gene Deletion Mutants: Strains with deletions in genes encoding porins (e.g., ΔompF), LPS biosynthesis enzymes (e.g., ΔwaaG), or chaperones (e.g., ΔsurA) are constructed. Increased susceptibility to specific antibiotics in these mutants confirms the role of these components in the permeability barrier [109].
- CRISPRi Depletion: The phenotype of a chemical inhibitor can be validated genetically. For example, the increased permeability caused by MAC13243 (a LolA inhibitor) was confirmed by using CRISPRi to knock down lolA expression, which recapitulated the NPN uptake and antibiotic sensitization phenotypes [109].
Table 2: Summary of Key Quantitative Assays for Compound Accumulation
| Assay Type | Measured Parameter | Key Strength(s) | Key Limitation(s) | Information Level |
|---|---|---|---|---|
| LC-MS/MS (Whole Cell) | Absolute intracellular concentration of parent compound. | Label-free; direct and quantitative; can be applied to any drug. | Labor-intensive; requires rapid separation; measures total cell accumulation, not subcellular localization. | Bulk, Quantitative |
| Extracellular Depletion MS | Rate of compound disappearance from medium. | Less disruptive; allows continuous monitoring. | Indirect measure; lower sensitivity for compounds with low accumulation. | Bulk, Quantitative |
| NPN Uptake | Relative outer membrane permeability. | Simple, high-throughput; excellent for identifying permeabilizers. | Indirect; does not measure the test antibiotic itself. | Bulk, Indirect |
| BLM Electrophysiology | Kinetics of single antibiotic molecules translocating through a specific porin. | Provides ultra-high resolution of translocation mechanism and kinetics. | Technically challenging; requires purified components; artificial membrane system. | Single-Molecule, Mechanistic |
| Synergy Checkerboard | Functional antimicrobial effect of drug combinations (FICI). | High clinical relevance; simple to perform. | Indirect measure of accumulation; does not provide mechanistic details. | Bulk, Functional |
| Deep UV Microscopy | Quantification of drug concentration within single cells. | Label-free; reveals population heterogeneity. | Limited to compounds with innate UV fluorescence. | Single-Cell, Quantitative |
Validating compound penetration through the Gram-negative cell envelope is a multifaceted challenge that requires a combination of sophisticated techniques. No single assay can provide a complete picture; rather, a synergistic approach is essential. Mass spectrometry offers direct quantification, fluorescence microscopy reveals spatial and temporal heterogeneity, electrophysiology deciphers molecular-scale interactions with porins, and microbiological synergy tests confirm functional relevance. By strategically integrating data from these biochemical, biophysical, and microbiological assays, researchers can robustly validate the penetration of novel compounds, guiding the rational design of effective antibiotics to combat the growing threat of multidrug-resistant Gram-negative infections.
The escalating global health crisis of antimicrobial resistance (AMR), particularly among Gram-negative bacteria, is driven by the formidable permeability barrier of the outer membrane. This membrane significantly limits the intracellular accumulation of antibiotics, contributing to high rates of treatment failure. Cefiderocol represents a paradigm shift in antibiotic design, directly addressing this challenge through a novel "Trojan horse" strategy that co-opts bacterial iron-uptake systems [111] [112]. Its development marks a critical advancement in the ongoing research to overcome outer membrane permeability and restore the efficacy of β-lactam antibiotics against multidrug-resistant pathogens.
Molecular Architecture and Siderophore Activity
Cefiderocol is a siderophore cephalosporin whose unique structure is central to its mechanism of action. Its molecular framework shares key features with established cephalosporins: a side chain at C-7 identical to that of ceftazidime, which confers stability against many β-lactamases and improves penetration, and a pyrrolidinium group at C-3 similar to cefepime, which enhances antibacterial potency and further improves β-lactamase stability [113] [112].
The critical innovation is the addition of a chlorocatechol group at the terminus of the C-3 side chain. This moiety mimics natural bacterial siderophores—high-affinity iron-chelating molecules like enterobactin and pyoverdine [113] [112]. At physiological pH, this chlorocatechol group chelates available ferric iron (Fe³⁺) to form a cefiderocol-Fe³⁺ complex [114]. This complex is actively transported across the outer membrane via specific bacterial iron transporters, such as CirA and Fiu in E. coli or PiuA in P. aeruginosa [112]. This active transport allows cefiderocol to bypass the passive diffusion limitations of porin channels and avoid efflux pump recognition, efficiently accumulating in the periplasmic space [112] [115].
Table 1: Key Structural Components of Cefiderocol and Their Functional Roles
| Structural Component | Chemical Feature | Functional Role |
|---|---|---|
| Cephalosporin Core | β-lactam ring | Binds to Penicillin-Binding Proteins (PBPs), primarily PBP-3, inhibiting bacterial cell wall synthesis [112]. |
| C-7 Side Chain | Identical to ceftazidime | Enhances stability against many β-lactamases and improves penetration across the outer membrane [112] [116]. |
| C-3 Side Chain | Pyrrolidinium group (as in cefepime) | Confers potent antibacterial activity and greater stability against β-lactamases [112]. |
| C-3 Terminus | Chlorocatechol group | Confers siderophore activity; chelates Fe³⁺ for active transport via iron transporters, bypassing porin and efflux-based resistance [113] [112]. |
The diagram below illustrates this iron-dependent active transport mechanism.
Mechanisms of Resistance
Despite its innovative design, resistance to cefiderocol has been reported, typically involving the convergence of multiple mechanisms [113]. Understanding these pathways is crucial for surveillance and preserving the drug's utility.
Key Resistance Pathways
The primary resistance mechanisms include:
Enzymatic Inactivation: Production of certain β-lactamases, particularly NDM-type metallo-β-lactamases (MBLs) and specific KPC variants (e.g., KPC-41), can hydrolyze or bind cefiderocol [113] [116]. Recent biochemical studies confirm that NDM-1 and NDM-5 efficiently hydrolyze cefiderocol, while other MBLs like VIM-2 and IMP-1 are inhibited by a stable enzyme-product adduct, explaining the strong clinical association between NDM production and cefiderocol resistance [116]. Other enzymes implicated include PER- and SHV-type extended-spectrum β-lactamases (ESBLs) [113].
Impaired Iron Transport: Mutations, truncations, or loss of function in genes encoding outer membrane siderophore receptors (e.g., cirA, fiu, piuA) directly compromise the active uptake of the cefiderocol-Fe³⁺ complex [113] [114]. Mutations in regulatory genes like envZ and tonB can also reduce the expression of these critical transporters [114].
Target Modification: Alterations in the target site, Penicillin-Binding Protein 3 (PBP-3), can reduce the binding affinity of cefiderocol, diminishing its antibacterial effect [113] [111].
Efflux Pumps and Porin Mutations: Although cefiderocol is not a substrate for most efflux pumps, some studies indicate efflux activity can contribute to resistance in some strains [114]. Mutations in porin genes can further reduce permeability, but this is often a secondary mechanism [114].
Table 2: Documented Resistance Mechanisms to Cefiderocol
| Resistance Mechanism | Molecular Basis | Key Examples |
|---|---|---|
| β-lactamase Production | Hydrolysis or binding of cefiderocol, preventing PBP binding. | NDM-type MBLs, KPC-41, KPC-50, PER-type, SHV-type ESBLs [113] [116]. |
| Iron Transporter Defects | Loss or mutation of siderophore receptors, preventing drug uptake. | Mutations in cirA, piuA, pirA; regulatory mutations in envZ, tonB [113] [114]. |
| Target Modification | Reduced drug binding to the target protein. | Mutations in PBP-3 [113] [111]. |
| Efflux & Porin Changes | Reduced intracellular concentration; often a contributory factor. | Efflux pump activity (inhibited by CCCP); porin mutations (OmpK35/36, OprD) [114]. |
The following diagram synthesizes these resistance pathways into a unified view.
Clinical and Real-World Evidence
Cefiderocol is indicated for complicated urinary tract infections (cUTIs), hospital-acquired bacterial pneumonia (HABP), and ventilator-associated bacterial pneumonia (VABP) caused by susceptible Gram-negative microorganisms [117]. Real-world evidence underscores its value in treating serious infections.
The international PROVE retrospective study analyzed over 1,000 patients in the U.S. and EU between 2020 and 2024. In the U.S. cohort (n=508), the overall clinical cure rate across different infection sites was 70.1% [118] [119]. Importantly, outcomes were significantly better when cefiderocol was used empirically (73.7% cure rate) compared to its use as salvage therapy (54.3%) [118] [119]. The drug demonstrated effectiveness against key pathogens including Pseudomonas aeruginosa (29.9% of monomicrobial infections), Acinetobacter baumannii (21.7%), and Stenotrophomonas maltophilia (4.9%) [118].
Data from the SENTRY Antimicrobial Surveillance Program (2020-2024) show that cefiderocol has maintained consistently high susceptibility rates among Gram-negative bacteria since its approval, including against carbapenem-resistant strains [118]. It also remains one of the few agents with activity against metallo-β-lactamase (MBL)-carrying A. baumannii [118]. Furthermore, about 90.6% of bacterial isolates non-susceptible to newer beta-lactam/beta-lactamase inhibitor combinations (BL-BLIs) remained susceptible to cefiderocol, demonstrating a low rate of cross-resistance [118].
Experimental Methodologies for Research
Studying cefiderocol's efficacy and resistance requires specific, standardized methodologies. The following outlines key experimental protocols used in recent research.
Susceptibility Testing and Genomic Analysis
A recent study investigating cefiderocol's activity against 370 clinical carbapenem-resistant Gram-negative bacteria (CRGNB) isolates employed the following workflow [114]:
Detailed Protocols:
Susceptibility Testing in ID-CAMHB: Susceptibility testing must be performed using iron-depleted cation-adjusted Mueller-Hinton broth (ID-CAMHB) to induce bacterial expression of iron-transport systems, a condition essential for cefiderocol's activity. The CLSI-recommended method involves treating MH medium with Chelex 100 resin to reduce iron concentration to <0.03 μg/mL, followed by supplementation with necessary cations (Zn²⁺, Ca²⁺, Mg²⁺) and filter sterilization [114]. Minimum Inhibitory Concentration (MIC) is then determined via microbroth dilution in this medium [114].
Efflux Pump Inhibition Assay: To investigate the role of efflux pumps in resistance, MIC determination is repeated in the presence of an efflux pump inhibitor such as Carbonyl Cyanide m-Chlorophenyl Hydrazone (CCCP). A significant reduction (e.g., ≥4-fold decrease) in the MIC value in the presence of CCCP indicates that active efflux contributes to the observed resistance phenotype [114].
Genomic Analysis of Resistance Mechanisms:
- Whole-Genome Sequencing (WGS): Resistant and susceptible isolates are subjected to WGS to identify mutations in genes associated with resistance, including those encoding siderophore receptors (cirA, piuA), porins, PBPs, and β-lactamases [114].
- RT-qPCR: Quantitative reverse transcription PCR is used to compare the expression levels of critical genes (e.g., iron transporters, β-lactamase genes, efflux pump components) between resistant and susceptible strains, identifying up- or down-regulation linked to the resistance phenotype [114].
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Studying Cefiderocol
| Research Reagent | Function in Experimental Protocol |
|---|---|
| Iron-Depleted Cation-Adjusted Mueller-Hinton Broth (ID-CAMHB) | The standardized culture medium for susceptibility testing; iron depletion stresses bacterial iron-acquisition systems, ensuring expression of transporters used by cefiderocol [114]. |
| Chelex 100 Resin | A chelating resin used to remove iron and other metal cations from Mueller-Hinton broth during the preparation of ID-CAMHB [114]. |
| Carbonyl Cyanide m-Chlorophenyl Hydrazone (CCCP) | An efflux pump inhibitor used in mechanistic studies to determine if reduced intracellular drug concentration contributes to resistance [114]. |
| Specific Primers/Probes for WGS & RT-qPCR | Targeted primers and probes are essential for sequencing and quantifying the expression of genes involved in resistance (e.g., blaNDM, cirA, piuA, ompK36, envZ) [114]. |
Cefiderocol is a groundbreaking antibiotic that successfully addresses the fundamental challenge of outer membrane permeability in Gram-negative bacteria. Its siderophore-based "Trojan horse" mechanism enables efficient penetration and accumulation in the periplasm, making it a potent therapeutic option against multidrug-resistant pathogens, including carbapenem-resistant strains. However, the emergence of resistance, particularly driven by NDM metallo-β-lactamases and mutations in iron transport systems, underscores the relentless adaptability of bacteria. Its continued efficacy will depend on the implementation of robust susceptibility testing, adherence to antimicrobial stewardship principles that favor earlier, empiric use in high-risk patients, and ongoing surveillance to monitor the evolution and spread of resistance mechanisms.
Conclusion
The outer membrane remains a critical determinant of antibiotic resistance in Gram-negative bacteria, functioning through a complex interplay of lipid asymmetry, porin-mediated diffusion, and synergistic efflux. Overcoming this barrier requires a multifaceted strategy that moves beyond traditional drug discovery. Future success hinges on the rational design of compounds optimized for specific porin pathways, the innovative use of active transport systems, and the strategic deployment of permeability enhancers and efflux pump inhibitors in combination therapies. Advancing tools to accurately measure compound penetration and a deeper, comparative understanding of pathogen-specific outer membrane architectures will be essential for translating these strategies into novel, effective treatments against the most urgent multidrug-resistant threats.
References
- 1. The asymmetric plasma membrane—A composite material ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11475564/]
- 2. Review Exploring membrane asymmetry and its effects on ... [https://www.sciencedirect.com/science/article/pii/S0968000424000215]
- 3. Outer membrane permeability and antibiotic resistance [https://pubmed.ncbi.nlm.nih.gov/19100346/]
- 4. Understanding antibiotic resistance via outer membrane ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5903844/]
- 5. Outer membrane permeability and antibiotic resistance [https://www.sciencedirect.com/science/article/abs/pii/S1570963908003592]
- 6. Building Asymmetric Lipid Bilayers for Molecular Dynamics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10384462/]
- 7. Quantitative imaging assessment of blood-brain barrier ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3570379/]
- 8. Function and Biogenesis of Lipopolysaccharides - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6091223/]
- 9. Biochemistry, Lipopolysaccharide - StatPearls - NCBI Bookshelf [https://www.ncbi.nlm.nih.gov/books/NBK554414/]
- 10. Outer Membrane Permeability and Antibiotic Resistance - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2696358/]
- 11. Lipopolysaccharide [https://en.wikipedia.org/wiki/Lipopolysaccharide]
- 12. Lipopolysaccharide: Recent Advances in Its Biosynthesis and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12387048/]
- 13. Bacterial lipopolysaccharide core structures mediate ... [https://www.sciencedirect.com/science/article/pii/S0005273619302986]
- 14. LPS O-antigen polysaccharide length impacts outer ... [https://pubmed.ncbi.nlm.nih.gov/41190655/]
- 15. LPS structure shapes T reg cells [https://www.nature.com/articles/s41590-025-02272-x]
- 16. Essential Roles of Hydrophobic Residues in Both MD-2 ... [https://www.sciencedirect.com/science/article/pii/S0021925819821166]
- 17. Role of Porins and Efflux Pumps in Drug Resistance - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3485749/]
- 18. Development of polymyxin- and aminoglycoside-based ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1625300/full]
- 19. Structure and Role of Gram-Negative Bacterial Porins - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3706956/]
- 20. Distinct Roles of Outer Membrane Porins in Antibiotic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6503746/]
- 21. Porin (protein) [https://grokipedia.com/page/Porin_(protein)]
- 22. Porin - an overview [https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/porin]
- 23. Structure and functional mechanism of porins - PubMed - NIH [https://pubmed.ncbi.nlm.nih.gov/8874494/]
- 24. Metabolic control of porin permeability influences antibiotic ... [https://www.nature.com/articles/s41564-025-02175-5]
- 25. Antimicrobial Resistance Facts and Stats [https://www.cdc.gov/antimicrobial-resistance/data-research/facts-stats/index.html]
- 26. Tackling the outer membrane: facilitating compound entry ... [https://www.nature.com/articles/s44259-023-00016-1]
- 27. Structure, Function and Regulation of Outer Membrane ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3617542/]
- 28. Impeding pathways of intrinsic resistance in Escherichia coli ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12543169/]
- 29. Impeding pathways of intrinsic resistance in Escherichia coli ... [https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3003443]
- 30. Antibiotic susceptibility signatures identify potential ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7481262/]
- 31. The Comprehensive Antibiotic Resistance Database [https://card.mcmaster.ca/]
- 32. Permeabilize, but Choose Wisely: Selective Antibiotic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12563807/]
- 33. New understanding of multi-drug efflux and permeation in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9839546/]
- 34. Types and Mechanisms of Efflux Systems and the Potential of... Pump [https://pmc.ncbi.nlm.nih.gov/articles/PMC10974122/]
- 35. - Wikipedia Efflux pump [https://en.wikipedia.org/wiki/Efflux_pump]
- 36. Multidrug efflux : pumps ... | Nature Reviews Microbiology structure [https://www.nature.com/articles/s41579-018-0048-6?error=cookies_not_supported&code=e40a9db3-7a2d-407c-ad2c-c38ebef3c666]
- 37. Efflux pumps and membrane permeability contribute to ... [https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1013027]
- 38. Sequence- and structure-based computational analyses of ... [https://www.sciencedirect.com/science/article/pii/S092325081830010X]
- 39. Predicting permeation of compounds across the outer ... [https://www.nature.com/articles/s42004-024-01161-y]
- 40. The Influence of Permeability through Bacterial Porins in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8226570/]
- 41. The Influence of Permeability through Bacterial Porins in ... [https://www.mdpi.com/2079-6382/10/6/635]
- 42. Seeking Correlation Among Porin Permeabilities and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11944608/]
- 43. Fast prediction of antibiotic permeability through ... [https://www.sciencedirect.com/science/article/pii/S0006349523002084]
- 44. Active transport of siderophore-mimicking antibacterials ... [https://www.sciencedirect.com/science/article/pii/S1368764699901073]
- 45. Unraveling the mechanisms behind the enhanced efficacy ... [https://www.nature.com/articles/s42003-025-08898-9]
- 46. Recent Advances in Strategies to Combat Bacterial Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10141387/]
- 47. Ferroptosis-new avenues for the treatment of multidrug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12537265/]
- 48. Siderophores as tools and treatments [https://www.nature.com/articles/s44259-024-00053-4]
- 49. Siderophore–Antibiotic Conjugate Design: New Drugs for ... [https://www.mdpi.com/1420-3049/24/18/3314]
- 50. Siderophore conjugates to combat antibiotic-resistant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10211321/]
- 51. Antibiotic conjugates: Using molecular Trojan Horses to ... [https://www.sciencedirect.com/science/article/pii/S075333222500201X]
- 52. Tackling microbial iron homeostasis: novel antimicrobial ... [https://www.sciencedirect.com/science/article/pii/S0165614725001798]
- 53. the various mechanisms of action of antimicrobial peptides ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3856679/]
- 54. Interaction of designed cationic antimicrobial peptides with ... [https://www.nature.com/articles/s41598-024-51716-1]
- 55. Interplay between porin deficiency, fitness, and virulence in ... [https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1012902]
- 56. Porin-Independent Uptake of Small Molecule Antibiotics ... [https://pubmed.ncbi.nlm.nih.gov/41042070/]
- 57. Exploring the permeation of fluoroquinolone ... [https://www.sciencedirect.com/science/article/pii/S0005273621002868]
- 58. Metabolic state-driven nutrient-based approach to combat ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11971349/]
- 59. Enhancing permeability of the outer membrane – by Helen ... [https://revive.gardp.org/enhancing-permeability-of-the-outer-membrane/]
- 60. Outer Membrane Disruption Overcomes Intrinsic, Acquired, ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7512548/]
- 61. AaeAP2a, a scorpion-derived antimicrobial peptide ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1673333/full]
- 62. Novel triple drug combination effective against antibiotic- ... [https://www.ox.ac.uk/news/2024-05-03-novel-triple-drug-combination-effective-against-antibiotic-resistant-bacteria]
- 63. Structure–activity relationship of a peptide permeation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9870020/]
- 64. Interplay between porin deficiency, fitness, and virulence in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11805372/]
- 65. implications for the introduction of imipenem-relebactam [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1689777/full]
- 66. Outer Membrane Porins Contribute to Antimicrobial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10385648/]
- 67. Altered Antibiotic Transport in OmpC Mutants Isolated from a ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0025825]
- 68. Porin loss in Klebsiella pneumoniae clinical isolates ... [https://www.sciencedirect.com/science/article/abs/pii/S1438422118303242]
- 69. Porin expression in clinical isolates of Klebsiella pneumoniae ... [https://ann-clinmicrob.biomedcentral.com/articles/10.1186/s12941-024-00761-9]
- 70. Role and mechanism of the outer membrane porin LamB in ... [https://www.sciencedirect.com/science/article/abs/pii/S0304389424030164]
- 71. A new antibiotic traps lipopolysaccharide in its ... [https://www.nature.com/articles/s41586-023-06799-7]
- 72. Characterization of Lipid A Modifications [https://www.dental.umaryland.edu/micropath/research/dr-robert-ernst-lab/projects/characterization-of-lipid-a-modifications-/]
- 73. Lipopolysaccharide as an antibiotic target [https://www.sciencedirect.com/science/article/pii/S0167488923000794]
- 74. lipid A structural modification systems offer diagnostic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10549895/]
- 75. Molecular Mechanisms of Bacterial Resistance to ... [https://www.mdpi.com/2076-2607/12/7/1259]
- 76. Induction of lipid A modification genes in Pseudomonas ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12494483/]
- 77. Outer Membrane Vesicle Production Facilitates LPS ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5082901/]
- 78. Phage Therapy as a Novel Alternative to Antibiotics ... [https://www.mdpi.com/2079-6382/14/10/1040]
- 79. Antibiotic Resistance: A Genetic and Physiological Perspective [https://pmc.ncbi.nlm.nih.gov/articles/PMC12579171/]
- 80. Beyond the Resistome: Molecular Insights, Emerging ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12561093/]
- 81. Understanding antibiotic resistance via outer membrane permeability [https://www.dovepress.com/understanding-antibiotic-resistance-via-outer-membrane-permeability-peer-reviewed-fulltext-article-IDR]
- 82. Role of bacterial efflux pumps in antibiotic resistance ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10268834/]
- 83. A permeability-increasing drug synergizes with bacterial ... [https://www.nature.com/articles/s41598-019-39659-4]
- 84. Inhibition of efflux pumps by FDA-approved drugs ... [https://pubmed.ncbi.nlm.nih.gov/40757818/]
- 85. Efflux pump inhibitors from plant compounds: A new ... [https://www.sciencedirect.com/science/article/pii/S0926669025009240]
- 86. Bac-EPIC: A web interface for developing novel efflux ... [https://www.sciencedirect.com/science/article/pii/S2590098623000143]
- 87. WHO warns of widespread resistance to common ... [https://www.who.int/news/item/13-10-2025-who-warns-of-widespread-resistance-to-common-antibiotics-worldwide]
- 88. Large-scale combination screens reveal small-molecule ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12002207/]
- 89. Synergistic combinations of antimicrobial peptides and ... [https://www.sciencedirect.com/science/article/pii/S0031699725075131]
- 90. Flash optimization of drug combinations for Acinetobacter ... [https://www.nature.com/articles/s44259-025-00079-2]
- 91. A Lack of Synergy Between Membrane-permeabilizing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4259837/]
- 92. Outer membrane permeability in Pseudomonas aeruginosa [https://pubmed.ncbi.nlm.nih.gov/6803666/]
- 93. Structural Insights into Outer of... Membrane Permeability [https://pubmed.ncbi.nlm.nih.gov/26805524/]
- 94. Contemporaneous Measurement of Outer and Inner Membrane ... [https://pubmed.ncbi.nlm.nih.gov/33659522/]
- 95. Antibiotic resistance in Pseudomonas aeruginosa [https://www.sciencedirect.com/science/article/pii/S0734975018301976]
- 96. Frontiers | Outer of mcr-positive bacteria... membrane permeability [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2024.1468682/full]
- 97. Resistance mechanisms and therapeutic strategies for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12576158/]
- 98. Structure, Dynamics, and Substrate Specificity of the OprO ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4601001/]
- 99. Structure and function of OprD protein in Pseudomonas ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3831278/]
- 100. Loss of OprD function is sufficient for carbapenem-resistance ... [https://bmcmicrobiol.biomedcentral.com/articles/10.1186/s12866-025-03935-3]
- 101. new evidence on the role of OprD and OpdP porins ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11960084/]
- 102. Reinforcing Lipid A Acylation on the Cell Surface of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4442142/]
- 103. Synthesis of Acinetobacter baumannii Lipid A(s) and ... [https://www.nature.com/articles/s42004-025-01628-6]
- 104. 2-Hydroxylation of Acinetobacter baumannii Lipid A ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6434125/]
- 105. Bilayer Properties of Lipid A from Various Gram-Negative ... [https://www.sciencedirect.com/science/article/pii/S0006349516307615]
- 106. Structural Insights into Outer Membrane Permeability of ... [https://www.sciencedirect.com/science/article/pii/S0969212615005353]
- 107. Quantifying Antibiotic Permeability across Gram-negative ... [https://www.sciencedirect.com/science/article/abs/pii/S0022283619301810]
- 108. Outer Membrane Porins Contribute to Antimicrobial ... [https://www.mdpi.com/2076-2607/11/7/1690]
- 109. Increasing the permeability of Escherichia coli using ... [https://www.nature.com/articles/s41598-017-17772-6]
- 110. A Barrier to Entry: Examining the Bacterial Outer ... [https://www.mdpi.com/2076-3417/13/7/4238]
- 111. Addressing antimicrobial resistance: Structural insights into ... [https://www.sciencedirect.com/science/article/pii/S1876034125002205]
- 112. Mechanism of action of cefiderocol - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9632059/]
- 113. Cefiderocol: Systematic Review of Mechanisms ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9220290/]
- 114. In vitro antimicrobial activity and resistance mechanisms of ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1670179/full]
- 115. Mechanism of action of cefiderocol [https://pubmed.ncbi.nlm.nih.gov/36193980/]
- 116. Cefiderocol “under siege”? Understanding the rise of NDM ... [https://pubs.rsc.org/en/content/articlehtml/2025/sc/d5sc02122g]
- 117. Fetroja® (cefiderocol) | Structure [https://www.fetroja.com/overcoming-carbapenem-resistance]
- 118. IDWeek 2025: Shionogi Presents Real-World and ... [https://www.shionogi.com/global/en/news/2025/10/20251021.html]
- 119. Drugmakers announce encouraging new antibiotic data at ... [https://www.cidrap.umn.edu/antimicrobial-stewardship/drugmakers-announce-encouraging-new-antibiotic-data-idweek]